Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Tyrosine-protein phosphatase non-receptor type 1 [1-298]
Ligand
BDBM13597
Substrate
BDBM13594
Meas. Tech.
PTP1B and TCPTP Inhibition Assay
pH
6.3±n/a
Temperature
295.15±n/a K
IC50
23±n/a nM
Citation
 Scapin, G; Patel, SB; Becker, JW; Wang, Q; Desponts, C; Waddleton, D; Skorey, K; Cromlish, W; Bayly, C; Therien, M; Gauthier, JY; Li, CS; Lau, CK; Ramachandran, C; Kennedy, BP; Asante-Appiah, E The structural basis for the selectivity of benzotriazole inhibitors of PTP1B. Biochemistry 42:11451-9 (2003) [PubMed] Article
Scapin, G; Patel, SB; Becker, JW; Wang, Q; Desponts, C; Waddleton, D; Skorey, K; Cromlish, W; Bayly, C; Therien, M; Gauthier, JY; Li, CS; Lau, CK; Ramachandran, C; Kennedy, BP; Asante-Appiah, E The structural basis for the selectivity of benzotriazole inhibitors of PTP1B. Biochemistry 42:11451-9 (2003) [PubMed] Article More Info.:
Target
Name:
Tyrosine-protein phosphatase non-receptor type 1 [1-298]
Synonyms:
PTN1_HUMAN | PTP-1B | PTP1B | PTPN1 | PTPase 1B | Protein-Tyrosine Phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase, non-receptor type 1
Type:
Enzyme
Mol. Mass.:
34670.65
Organism:
Homo sapiens (Human)
Description:
The catalytic domain of PTP 1B (residues 1-298) was expressed and purified from E. coli.
Residue:
298
Sequence:
MEMEKEFEQIDKSGSWAAIYQDIRHEASDFPCRVAKLPKNKNRNRYRDVSPFDHSRIKLHQEDNDYINASLIKMEEAQRSYILTQGPLPNTCGHFWEMVWEQKSRGVVMLNRVMEKGSLKCAQYWPQKEEKEMIFEDTNLKLTLISEDIKSYYTVRQLELENLTTQETREILHFHYTTWPDFGVPESPASFLNFLFKVRESGSLSPEHGPVVVHCSAGIGRSGTFCLADTCLLLMDKRKDPSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHED
Inhibitor
Name:
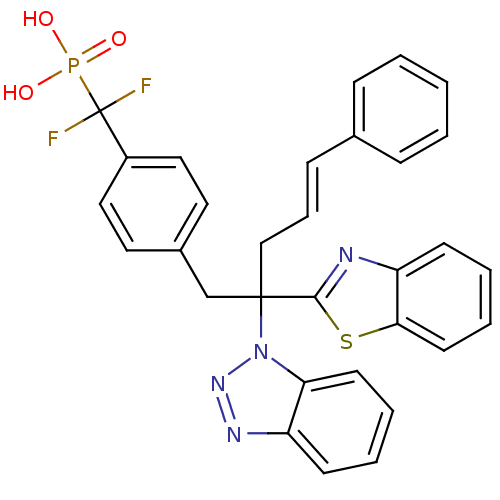
BDBM13597
Synonyms:
({4-[(4E)-2-(1,3-benzothiazol-2-yl)-2-(1H-1,2,3-benzotriazol-1-yl)-5-phenylpent-4-en-1-yl]phenyl}difluoromethyl)phosphonic acid | Aryldifluoromethyl-Phosphonic Acid Inhibitor 4 | [[4-[(E,2R)-2-benzothiazol-2-yl-2-benzotriazol-1-yl-5-phenyl-pent-4-enyl]phenyl]-difluoro-methyl]phosphonic acid
Type:
Small organic molecule
Emp. Form.:
C31H25F2N4O3PS
Mol. Mass.:
602.591
SMILES:
OP(O)(=O)C(F)(F)c1ccc(CC(C\C=C\c2ccccc2)(c2nc3ccccc3s2)n2nnc3ccccc23)cc1
Substrate
Name:
BDBM13594
Synonyms:
Fluorescein diphosphate (FDP) | ammonium 6'-(phosphonatooxy)-3H-spiro[2-benzofuran-1,9'-xanthene]-3-one phosphate | fluorescein diphosphate, tetraammonium salt
Type:
Small organic molecule
Emp. Form.:
C20H10O11P2
Mol. Mass.:
488.2365
SMILES:
[O-]P([O-])(=O)Oc1ccc2c(Oc3cc(OP([O-])([O-])=O)ccc3C22OC(=O)c3ccccc23)c1