Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Sodium-dependent neutral amino acid transporter B(0)AT2
Ligand
BDBM50061421
Substrate
n/a
Meas. Tech.
ChEMBL_1432515 (CHEMBL3386947)
IC50
16100±n/a nM
Citation
 Cuboni, S; Devigny, C; Hoogeland, B; Strasser, A; Pomplun, S; Hauger, B; Höfner, G; Wanner, KT; Eder, M; Buschauer, A; Holsboer, F; Hausch, F Loratadine and analogues: discovery and preliminary structure-activity relationship of inhibitors of the amino acid transporter B(0)AT2. J Med Chem 57:9473-9 (2014) [PubMed] Article
Cuboni, S; Devigny, C; Hoogeland, B; Strasser, A; Pomplun, S; Hauger, B; Höfner, G; Wanner, KT; Eder, M; Buschauer, A; Holsboer, F; Hausch, F Loratadine and analogues: discovery and preliminary structure-activity relationship of inhibitors of the amino acid transporter B(0)AT2. J Med Chem 57:9473-9 (2014) [PubMed] Article More Info.:
Target
Name:
Sodium-dependent neutral amino acid transporter B(0)AT2
Synonyms:
B0AT2 | B0AT2 | NTT73 | S6A15_HUMAN | SBAT1 | SLC6A15 | Sodium- and chloride-dependent neurotransmitter transporter NTT73 | Sodium-coupled branched-chain amino-acid transporter 1 | Sodium-dependent neutral amino acid transporter B(0)AT2 | Solute carrier family 6 member 15 | Transporter v7-3
Type:
PROTEIN
Mol. Mass.:
81820.70
Organism:
Homo sapiens (Human)
Description:
ChEMBL_109513
Residue:
730
Sequence:
MPKNSKVVKRELDDDVTESVKDLLSNEDAADDAFKTSELIVDGQEEKDTDVEEGSEVEDERPAWNSKLQYILAQVGFSVGLGNVWRFPYLCQKNGGGAYLLPYLILLMVIGIPLFFLELSVGQRIRRGSIGVWNYISPKLGGIGFASCVVCYFVALYYNVIIGWSLFYFSQSFQQPLPWDQCPLVKNASHTFVEPECEQSSATTYYWYREALNISSSISESGGLNWKMTICLLAAWVMVCLAMIKGIQSSGKIIYFSSLFPYVVLICFLIRAFLLNGSIDGIRHMFTPKLEIMLEPKVWREAATQVFFALGLGFGGVIAFSSYNKRDNNCHFDAVLVSFINFFTSVLATLVVFAVLGFKANVINEKCITQNSETIMKFLKMGNISQDIIPHHINLSTVTAEDYHLVYDIIQKVKEEEFPALHLNSCKIEEELNKAVQGTGLAFIAFTEAMTHFPASPFWSVMFFLMLVNLGLGSMFGTIEGIVTPIVDTFKVRKEILTVICCLLAFCIGLIFVQRSGNYFVTMFDDYSATLPLLIVVILENIAVCFVYGIDKFMEDLKDMLGFAPSRYYYYMWKYISPLMLLSLLIASVVNMGLSPPGYNAWIEDKASEEFLSYPTWGLVVCVSLVVFAILPVPVVFIVRRFNLIDDSSGNLASVTYKRGRVLKEPVNLEGDDTSLIHGKIPSEMPSPNFGKNIYRKQSGSPTLDTAPNGRYGIGYLMADIMPDMPESDL
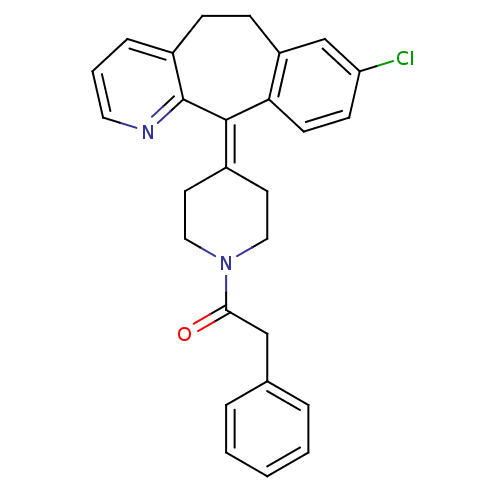
Inhibitor
Name:
BDBM50061421
Synonyms:
1-[4-(8-Chloro-5,6-dihydro-benzo[5,6]cyclohepta[1,2-b]pyridin-11-ylidene)-piperidin-1-yl]-2-phenyl-ethanone | CHEMBL338697
Type:
Small organic molecule
Emp. Form.:
C27H25ClN2O
Mol. Mass.:
428.953
SMILES:
Clc1ccc2c(-[#6]-[#6]-c3cccnc3\[#6]-2=[#6]-2/[#6]-[#6]-[#7](-[#6]-[#6]-2)-[#6](=O)-[#6]-c2ccccc2)c1