Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Voltage-dependent L-type calcium channel subunit alpha-1C
Ligand
BDBM50031720
Substrate
n/a
Meas. Tech.
ChEMBL_1479100 (CHEMBL3436076)
IC50
1023000±n/a nM
Citation
 Wisniowska, B; Mendyk, A; Fijorek, K; Glinka, A; Polak, S Predictive model for L-type channel inhibition: multichannel block in QT prolongation risk assessment. J Appl Toxicol 32:858-66 (2012) [PubMed] Article
Wisniowska, B; Mendyk, A; Fijorek, K; Glinka, A; Polak, S Predictive model for L-type channel inhibition: multichannel block in QT prolongation risk assessment. J Appl Toxicol 32:858-66 (2012) [PubMed] Article More Info.:
Target
Name:
Voltage-dependent L-type calcium channel subunit alpha-1C
Synonyms:
CAC1C_HUMAN | CACH2 | CACN2 | CACNA1C | CACNL1A1 | CCHL1A1 | Calcium channel (Type L) | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | L-type calcium channel alpha-1c/beta-2/alpha2delta-1 | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav1.2
Type:
Enzyme Catalytic Domain
Mol. Mass.:
248979.79
Organism:
Homo sapiens (Human)
Description:
Calcium channel (Type L) 0 HUMAN::Q13936
Residue:
2221
Sequence:
MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPKKQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIAYGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLHIALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCITMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPENEDEGMDEEKPRNMSMPTSETESVNTENVAGGDIEGENCGARLAHRISKSKFSRYWRRWNRFCRRKCRAAVKSNVFYWLVIFLVFLNTLTIASEHYNQPNWLTEVQDTANKALLALFTAEMLLKMYSLGLQAYFVSLFNRFDCFVVCGGILETILVETKIMSPLGISVLRCVRLLRIFKITRYWNSLSNLVASLLNSVRSIASLLLLLFLFIIIFSLLGMQLFGGKFNFDEMQTRRSTFDNFPQSLLTVFQILTGEDWNSVMYDGIMAYGGPSFPGMLVCIYFIILFICGNYILLNVFLAIAVDNLADAESLTSAQKEEEEEKERKKLARTASPEKKQELVEKPAVGESKEEKIELKSITADGESPPATKINMDDLQPNENEDKSPYPNPETTGEEDEEEPEMPVGPRPRPLSELHLKEKAVPMPEASAFFIFSSNNRFRLQCHRIVNDTIFTNLILFFILLSSISLAAEDPVQHTSFRNHILFYFDIVFTTIFTIEIALKILGNADYVFTSIFTLEIILKMTAYGAFLHKGSFCRNYFNILDLLVVSVSLISFGIQSSAINVVKILRVLRVLRPLRAINRAKGLKHVVQCVFVAIRTIGNIVIVTTLLQFMFACIGVQLFKGKLYTCSDSSKQTEAECKGNYITYKDGEVDHPIIQPRSWENSKFDFDNVLAAMMALFTVSTFEGWPELLYRSIDSHTEDKGPIYNYRVEISIFFIIYIIIIAFFMMNIFVGFVIVTFQEQGEQEYKNCELDKNQRQCVEYALKARPLRRYIPKNQHQYKVWYVVNSTYFEYLMFVLILLNTICLAMQHYGQSCLFKIAMNILNMLFTGLFTVEMILKLIAFKPKGYFSDPWNVFDFLIVIGSIIDVILSETNHYFCDAWNTFDALIVVGSIVDIAITEVNPAEHTQCSPSMNAEENSRISITFFRLFRVMRLVKLLSRGEGIRTLLWTFIKSFQALPYVALLIVMLFFIYAVIGMQVFGKIALNDTTEINRNNNFQTFPQAVLLLFRCATGEAWQDIMLACMPGKKCAPESEPSNSTEGETPCGSSFAVFYFISFYMLCAFLIINLFVAVIMDNFDYLTRDWSILGPHHLDEFKRIWAEYDPEAKGRIKHLDVVTLLRRIQPPLGFGKLCPHRVACKRLVSMNMPLNSDGTVMFNATLFALVRTALRIKTEGNLEQANEELRAIIKKIWKRTSMKLLDQVVPPAGDDEVTVGKFYATFLIQEYFRKFKKRKEQGLVGKPSQRNALSLQAGLRTLHDIGPEIRRAISGDLTAEEELDKAMKEAVSAASEDDIFRRAGGLFGNHVSYYQSDGRSAFPQTFTTQRPLHINKAGSSQGDTESPSHEKLVDSTFTPSSYSSTGSNANINNANNTALGRLPRPAGYPSTVSTVEGHGPPLSPAIRVQEVAWKLSSNRERHVPMCEDLELRRDSGSAGTQAHCLLLRKANPSRCHSRESQAAMAGQEETSQDETYEVKMNHDTEACSEPSLLSTEMLSYQDDENRQLTLPEEDKRDIRQSPKRGFLRSASLGRRASFHLECLKRQKDRGGDISQKTVLPLHLVHHQALAVAGLSPLLQRSHSPASFPRPFATPPATPGSRGWPPQPVPTLRLEGVESSEKLNSSFPSIHCGSWAETTPGGGGSSAARRVRPVSLMVPSQAGAPGRQFHGSASSLVEAVLISEGLGQFAQDPKFIEVTTQELADACDMTIEEMESAADNILSGGAPQSPNGALLPFVNCRDAGQDRAGGEEDAGCVRARGRPSEEELQDSRVYVSSL
Inhibitor
Name:
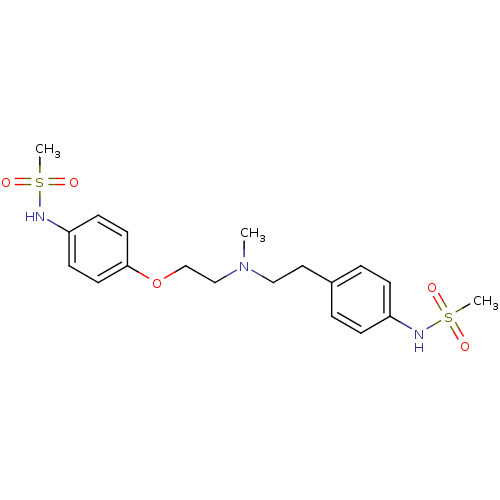
BDBM50031720
Synonyms:
(Dofetilide) N-[4-(2-{[2-(4-Methanesulfonylamino-phenyl)-ethyl]-methylamino}-ethoxy)-phenyl]-methanesulfonamide | CHEMBL473 | DOFETILIDE | N-[4-(2-{[2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-METHYL-AMINO}-ETHOXY)-PHENYL]-METHANESULFONAMIDE DOFETILIDE | N-[4-(2-{[2-(4-Methanesulfonylamino-phenyl)-ethyl]-methyl-amino}-ethoxy)-phenyl]-methanesulfonamide (UK-68798) | N-[4-(2-{[2-(4-Methanesulfonylamino-phenyl)-ethyl]-methyl-amino}-ethoxy)-phenyl]-methanesulfonamide (dofetilide) | N-[4-(2-{[2-(4-methanesulfonylamino-phenoxy)-ethyl]-methyl-amino}-ethyl)-phenyl]-methanesulfonamide | TIKOSYN | UK-68,798 | US10167299, Dofetilide
Type:
Small organic molecule
Emp. Form.:
C19H27N3O5S2
Mol. Mass.:
441.565
SMILES:
CN(CCOc1ccc(NS(C)(=O)=O)cc1)CCc1ccc(NS(C)(=O)=O)cc1