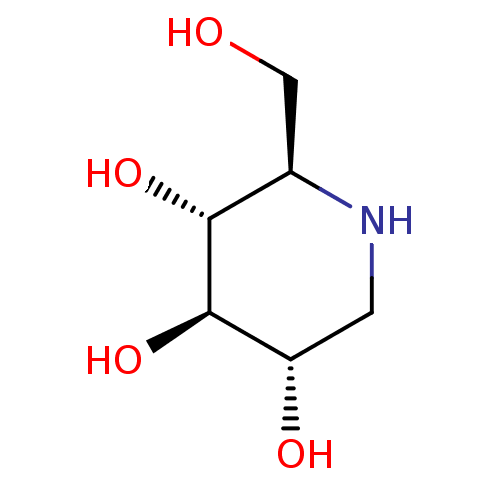
BDBM18351 (2R,3R,4R,5S)-2-(Hydroxymethyl)piperidine-3,4,5-triol, 10::(2R,3R,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol::1-Deoxynojirimycin::1-deoxynojirimycin (DNJ)::CHEMBL307429::US20230339856, Compound DNJ::US9181184, 1::dNM
SMILES OC[C@H]1NC[C@H](O)[C@@H](O)[C@@H]1O
InChI Key InChIKey=LXBIFEVIBLOUGU-JGWLITMVSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 18351
Found 3 hits for monomerid = 18351
Affinity DataIC50: 3.95E+4nMAssay Description:Inhibition of Saccharomyces cerevisiae alpha-glucosidase using PNPG as substrate incubated for 30 mins by spectrophotometric assayMore data for this Ligand-Target Pair
Affinity DataIC50: 6.88E+4nMAssay Description:Inhibition of Saccharomyces cerevisiae alpha-glucosidase using maltose as substrate incubated for 30 mins by spectrophotometric assayMore data for this Ligand-Target Pair
Affinity DataIC50: 2.64E+4nMAssay Description:Inhibition of baker's yeast alpha-glucosidase using PNP as substrate preincubated for 30 mins followed by sucrose addition and measured after 1 mins ...More data for this Ligand-Target Pair