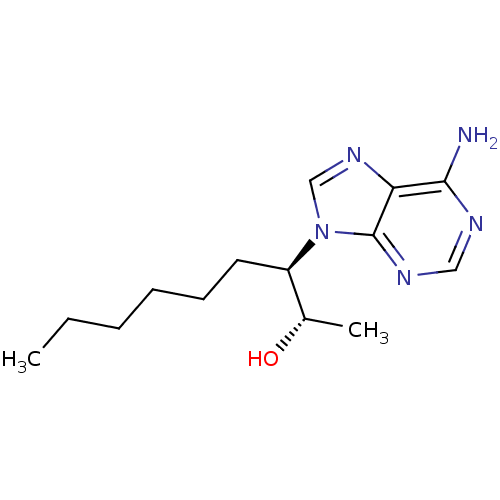
BDBM28393 (+)-EHNA::(2S,3R)-3-(6-amino-9H-purin-9-yl)nonan-2-ol::CHEMBL296435
SMILES CCCCCC[C@H]([C@H](C)O)n2cnc1c(N)ncnc12
InChI Key InChIKey=IOSAAWHGJUZBOG-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 26 hits for monomerid = 28393
Found 26 hits for monomerid = 28393
Affinity DataKi: 0.820nMAssay Description:Tested for binding affinity against calf intestinal Adenosine Deaminase (ADA)More data for this Ligand-Target Pair
Affinity DataKi: 1nMAssay Description:Binding affinity to ADA (unknown origin) assessed as inhibition constantMore data for this Ligand-Target Pair
Affinity DataKi: 1.10nMAssay Description:Inhibitory activity against adenosine deaminase in calf intestinal mucosa.More data for this Ligand-Target Pair
Affinity DataKi: 1.13nMAssay Description:Inhibition of Adenosine deaminase (ADA) of calf intestineMore data for this Ligand-Target Pair
Affinity DataKi: 1.14nMAssay Description:Inhibitory constant against bovine spleen Adenosine deaminaseMore data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Binding affinity (Ki) at calf intestinal Adenosine deaminaseMore data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Compound was evaluated for inhibition of calf intestinal mucosa adenosine deaminase (ADA)More data for this Ligand-Target Pair
Affinity DataKi: 2nMAssay Description:Compound was evaluated for the inhibition of adenosine deaminaseMore data for this Ligand-Target Pair
Affinity DataKi: 2.20nMAssay Description:Inhibition of bovine adenosine deaminase pre-incubated for 5 mins before adenosine additionMore data for this Ligand-Target Pair
Affinity DataIC50: 635nMAssay Description:Inhibitory activity against PDE2 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 800nMAssay Description:Inhibition of human PDE2 using c[8-3H]cAMP or c[8-3H]cGMP as substrate measured after 10 mins by liquid scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.14E+3nM ΔG°: -8.24kcal/molepH: 7.2 T: 2°CAssay Description:The activity of ADA was determined spectrophotometrically by monitoring the change in absorbance at 262 nm, due to the deamination of adenosine catal...More data for this Ligand-Target Pair
Affinity DataIC50: 2.46E+3nMAssay Description:Inhibition of recombinant human PDE2A catalytic domain (580 to 919 residues) expressed in Escherichia coli BL21 (Codonplus) using [3H]cGMP as substra...More data for this Ligand-Target Pair
Affinity DataIC50: 2.62E+3nMAssay Description:Inhibition of human recombinant PDE2A using cGMP as substrate incubated for 20 mins measured by Kinase Glo reagent based microplate reader assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE9 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE10 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE4C by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE11 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE3 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE4B by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE4D by scintillation proximity assayMore data for this Ligand-Target Pair
TargetHigh affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B(Human)
Pfizer
Curated by ChEMBL
Pfizer
Curated by ChEMBL
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE8B by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE4A by scintillation proximity assayMore data for this Ligand-Target Pair
TargetDual specificity calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A/1B/1C(Human)
Pfizer
Curated by ChEMBL
Pfizer
Curated by ChEMBL
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE1 by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE5 by scintillation proximity assayMore data for this Ligand-Target Pair
TargetHigh affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A(Human)
Pfizer
Curated by ChEMBL
Pfizer
Curated by ChEMBL
Affinity DataIC50: 1.60E+4nMAssay Description:Inhibitory activity against PDE8A by scintillation proximity assayMore data for this Ligand-Target Pair