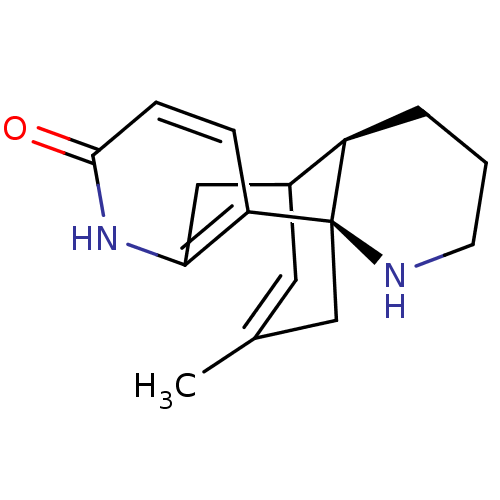
BDBM10632 (-)-Huperzine B::(1R,10R)-16-methyl-6,14-diazatetracyclo[7.5.3.0^{1,10}.0^{2,7}]heptadeca-2(7),3,16-trien-5-one
SMILES CC1=CC2Cc3[nH]c(=O)ccc3[C@]3(C1)NCCC[C@H]23
InChI Key InChIKey=YYWGABLTRMRUIT-ZLXOECBPSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 10632
Found 3 hits for monomerid = 10632
Affinity DataKi: 334nM ΔG°: -8.83kcal/molepH: 7.0 T: 25°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.93E+4nMpH: 7.4 T: 37°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at...More data for this Ligand-Target Pair
Affinity DataIC50: 2.27E+5nMpH: 7.4 T: 37°CAssay Description:The cholinesterase assays were performed using colorimetric method reported by Ellman. Enzyme activity was determined by measuring the absorbance at...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)