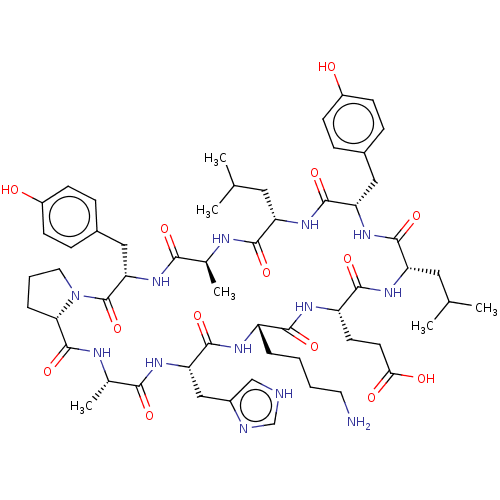
BDBM50626274 CHEMBL5429947
SMILES [H][C@@]12CCCN1C(=O)[C@H](Cc1ccc(O)cc1)NC(=O)[C@H](C)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](Cc1ccc(O)cc1)NC(=O)[C@H](CC(C)C)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCCCN)NC(=O)[C@H](Cc1c[nH]cn1)NC(=O)[C@H](C)NC2=O
InChI Key InChIKey=PVHUBGNZLXZQSU-UHFFFAOYSA-N
Data 2 Kd
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50626274
Found 2 hits for monomerid = 50626274
TargetTranscriptional coactivator YAP1(Homo sapiens)
Shanghai Institute of Materia Medica
Curated by ChEMBL
Shanghai Institute of Materia Medica
Curated by ChEMBL
Affinity DataKd: 340nMAssay Description:Binding affinity to N-terminal YAP ( 1 to 291 residues) (unknown origin) expressed in Escherichia coli Rosetta cells assessed as dissociation constan...More data for this Ligand-Target Pair
TargetTranscriptional coactivator YAP1(Homo sapiens)
Shanghai Institute of Materia Medica
Curated by ChEMBL
Shanghai Institute of Materia Medica
Curated by ChEMBL
Affinity DataKd: 2.64E+3nMAssay Description:Binding affinity to YAP WW domain (165 to 271 residues) (unknown origin) expressed in Escherichia coli Rosetta cells assessed as dissociation constan...More data for this Ligand-Target Pair