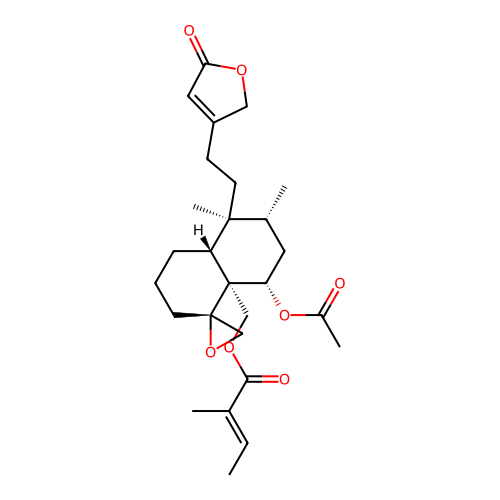
BDBM50646883 AJUGACUMBIN A
SMILES C/C=C(\C)C(=O)OC[C@@]12[C@@H](OC(C)=O)C[C@@H](C)[C@](C)(CCC3=CC(=O)OC3)[C@H]1CCC[C@]21CO1
InChI Key
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50646883
Found 3 hits for monomerid = 50646883
TargetPeroxisome proliferator-activated receptor gamma(Human)
Sun Yat-sen University
Curated by ChEMBL
Sun Yat-sen University
Curated by ChEMBL
Affinity DataIC50: 8.35E+3nMAssay Description:Antagonist activity at PPAR gamma LBD (203 to 476 residues) (unknown origin) expressed in HEK293T cells co-transfected with pCMV-GAL4-PPARgamma-LBD-S...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Human)
Sun Yat-sen University
Curated by ChEMBL
Sun Yat-sen University
Curated by ChEMBL
Affinity DataKd: 1.77E+4nMAssay Description:Binding affinity to PPAR gamma LBD (203 to 476 residues) (unknown origin) extracted from Escherichia coli BL21 (DE3) cells assessed as dissociation c...More data for this Ligand-Target Pair
TargetPeroxisome proliferator-activated receptor gamma(Human)
Sun Yat-sen University
Curated by ChEMBL
Sun Yat-sen University
Curated by ChEMBL
Affinity DataKd: 7.95E+4nMAssay Description:Binding affinity to PPAR gamma LBD (203 to 476 residues) (unknown origin) extracted from Escherichia coli BL21 (DE3) cells assessed as dissociation c...More data for this Ligand-Target Pair