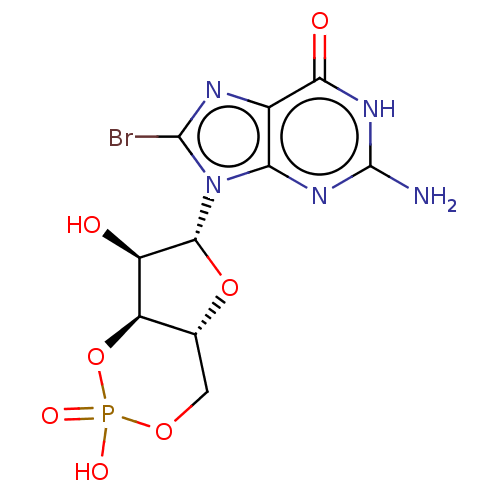
BDBM65502 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one::8-Br-cGMP
SMILES Nc1nc2n([C@@H]3O[C@@H]4COP(O)(=O)O[C@H]4[C@H]3O)c(Br)nc2c(=O)[nH]1
InChI Key InChIKey=YUFCOOWNNHGGOD-UMMCILCDSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 8 hits for monomerid = 65502
Found 8 hits for monomerid = 65502
Affinity DataEC50: 222nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
TargetIsoform Beta of cGMP-dependent protein kinase 1 [1-351,G182E](Human)
Baylor College of Medicine
Baylor College of Medicine
Affinity DataEC50: 765nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
TargetIsoform Beta of cGMP-dependent protein kinase 1 [1-351,G306E](Human)
Baylor College of Medicine
Baylor College of Medicine
Affinity DataEC50: <2nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
Affinity DataEC50: 30nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
Affinity DataEC50: 58nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
Affinity DataEC50: 20nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
Affinity DataEC50: 11nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
TargetIsoform Beta of cGMP-dependent protein kinase 1 [5-676,G306E](Human)
Baylor College of Medicine
Baylor College of Medicine
Affinity DataEC50: 206nMpH: 7.0Assay Description:Kinase activity was determined employing a microfluidic mobility-shift assay on a Caliper DeskTop Profiler (Caliper Life Sciences, PerkinElmer). The ...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)