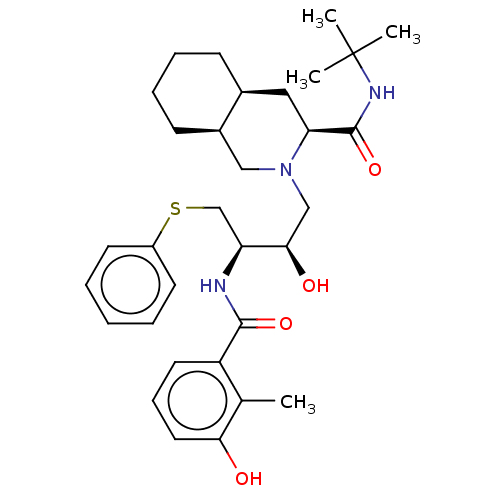
BDBM518 (3S,4aS,8aS)-N-tert-butyl-2-[(2R,3R)-2-hydroxy-3-[(3-hydroxy-2-methylphenyl)formamido]-4-(phenylsulfanyl)butyl]-decahydroisoquinoline-3-carboxamide::AG-1343::CHEMBL584::Nelfinavir::Viracept
SMILES Cc1c(O)cccc1C(=O)N[C@@H](CSc1ccccc1)[C@H](O)CN1C[C@H]2CCCC[C@H]2C[C@H]1C(=O)NC(C)(C)C
InChI Key InChIKey=QAGYKUNXZHXKMR-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 28 hits for monomerid = 518
Found 28 hits for monomerid = 518
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0100nM ΔG°: -15.0kcal/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,A572V](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 0.220nM ΔG°: -13.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,Q496K](Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
Affinity DataKi: 0.254nM ΔG°: -13.1kcal/molepH: 5.0 T: 2°CAssay Description:Protease activity was determined by following the increase in fluorescence with hydrolysis of the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Ty...More data for this Ligand-Target Pair
Affinity DataKi: 0.530nMpH: 5.5Assay Description:Protease inhibition fluorescence-based assay using cyclic ureas to inhibit HIV-protease.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,M537I,A572V](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 0.630nM ΔG°: -13.0kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,M537I](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 0.900nM ΔG°: -12.8kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,V583F](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 1.20nM ΔG°: -12.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 1.20nM ΔG°: -12.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 1.20nM ΔG°: -12.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 1.20nM ΔG°: -12.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,Q496K,I573V](Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
Affinity DataKi: 1.90nM ΔG°: -11.9kcal/molepH: 5.0 T: 2°CAssay Description:Protease activity was determined by following the increase in fluorescence with hydrolysis of the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Ty...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [482-580,I502K,I528M,T553A](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 2nM ΔG°: -12.3kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [482-580,I502K,I528M,T553A,V564F](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 3.70nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,D531N,M537I](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 6nM ΔG°: -11.7kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,D531N,A572V](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 6.10nM ΔG°: -11.6kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,D531N](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 6.80nM ΔG°: -11.6kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [490-588,L523I,E525D,M526I,I544V,L553H,H559K,L579M](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 17nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,L552P,A560V,V571A,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 17nM ΔG°: -11.0kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,I543V,L552P,A560V,V571A,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 23nM ΔG°: -10.8kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,L552P,A560V,V571A,I573V,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 24nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,D531N,M537I,A572V](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 26nM ΔG°: -10.8kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constant, Ki was determined by monitoring the competitive inhibition of the hydrolysis of the chromogenic substrate.More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,M535I,L552P,A560V,V571A,I573V,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 52nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,M535I,L552P,A560V,V571A,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 116nM ΔG°: -9.84kcal/molepH: 4.7 T: 2°CAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [490-588,L523I,E525D,M526I,I544V,L553H,H559K,V572F,L579M](Human immunodeficiency virus type 1)
Johnson & Johnson Pharmaceutical
Johnson & Johnson Pharmaceutical
Affinity DataKi: 146nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,Q496K,L499I,M525I,S526D,M535I,R546K,L552P,A560V,G562S,L579M,I582L](Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
Affinity DataKi: 445nM ΔG°: -8.66kcal/molepH: 5.0 T: 2°CAssay Description:Protease activity was determined by following the increase in fluorescence with hydrolysis of the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Ty...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,M535I,I543V,L552P,A560V,V571A,I573V,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 563nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,Q496K,L499I,M525I,S526D,M535I,R546K,L552P,A560V,G562S,I573V,L579M,I582L](Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
Affinity DataKi: 722nM ΔG°: -8.37kcal/molepH: 5.0 T: 2°CAssay Description:Protease activity was determined by following the increase in fluorescence with hydrolysis of the fluorogenic substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Ty...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [489-587,K509R,V521I,L522F,M525I,I543V,L552P,A560V,V571A,I573V,L579M](Human immunodeficiency virus type 1)
University of Florida College of Medicine
University of Florida College of Medicine
Affinity DataKi: 1.26E+3nMAssay Description:The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe...More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 18 hits for monomerid = 518
Found 18 hits for monomerid = 518
ITC DataΔG°: -11.4kcal/mole −TΔS°: -14.2kcal/mole ΔH°: 2.80kcal/mole
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease Mutant (V82F/I84V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -9.68kcal/mole −TΔS°: -14.1kcal/mole ΔH°: 4.39kcal/mole
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -12.8kcal/mole −TΔS°: -15.9kcal/mole ΔH°: 3.09kcal/mole logk: 2.20E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -9.58kcal/mole −TΔS°: -19.2kcal/mole ΔH°: 9.58kcal/mole logk: 1.10E+7
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -11.0kcal/mole −TΔS°: -18.3kcal/mole ΔH°: 7.29kcal/mole logk: 1.20E+8
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease Mutant (V82A/I84V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -12.4kcal/mole −TΔS°: -15.6kcal/mole ΔH°: 3.19kcal/mole logk: 1.20E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease Mutant (M46I/I54V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -12.4kcal/mole −TΔS°: -18.9kcal/mole ΔH°: 6.49kcal/mole logk: 1.20E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease Mutant (L10I/L90M)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -12.1kcal/mole −TΔS°: -16.9kcal/mole ΔH°: 4.79kcal/mole logk: 7.40E+8
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -13.1kcal/mole −TΔS°: -15.7kcal/mole ΔH°: 2.60kcal/mole logk: 3.80E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -12.6kcal/mole −TΔS°: -15.5kcal/mole ΔH°: 2.90kcal/mole logk: 1.69E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -12.5kcal/mole −TΔS°: -15.5kcal/mole ΔH°: 3.00kcal/mole logk: 1.41E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease B Subtype Mutant (V82F/I84V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -11.3kcal/mole −TΔS°: -15.7kcal/mole ΔH°: 4.39kcal/mole logk: 1.89E+8
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease C Subtype Mutant (V82F/I84V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -10.8kcal/mole −TΔS°: -15.0kcal/mole ΔH°: 4.19kcal/mole logk: 8.30E+7
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
CellHIV-1 Protease A Subtype Mutant (V82F/I84V)(Human immunodeficiency virus type 1)
The Johns Hopkins University
The Johns Hopkins University
ITC DataΔG°: -10.6kcal/mole −TΔS°: -14.7kcal/mole ΔH°: 4.09kcal/mole logk: 6.20E+7
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -8.29kcal/mole −TΔS°: -12.1kcal/mole ΔH°: 3.79kcal/mole logk: 1.17E+6
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -12.3kcal/mole −TΔS°: -17.1kcal/mole ΔH°: 4.79kcal/mole logk: 1.11E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -8.59kcal/mole −TΔS°: -14.2kcal/mole ΔH°: 5.59kcal/mole logk: 1.92E+6
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C
ITC DataΔG°: -13.1kcal/mole −TΔS°: -15.7kcal/mole ΔH°: 2.60kcal/mole logk: 3.85E+9
pH: 5.0 T: 25.00°C
pH: 5.0 T: 25.00°C