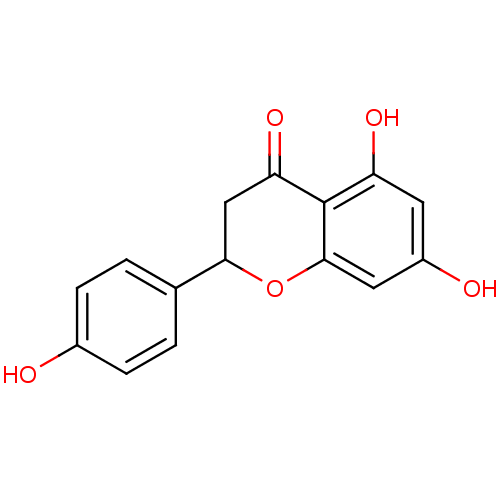
BDBM19461 α-CA inhibitor, 5::5,7-dihydroxy-2-(4-hydroxyphenyl)-3,4-dihydro-2H-1-benzopyran-4-one::CHEMBL32571::Naringenin::Naringenin (NAR)::naringetol::salipurpol
SMILES Oc1ccc(cc1)C1CC(=O)c2c(O)cc(O)cc2O1
InChI Key InChIKey=FTVWIRXFELQLPI-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 13 hits for monomerid = 19461
Found 13 hits for monomerid = 19461
Affinity DataKi: 0.00110nM IC50: 1.00E+3nMAssay Description:Inhibition assay using aromatase enzyme.More data for this Ligand-Target Pair
Affinity DataKi: 113nMpH: 7.4Assay Description:CA activity was assayed by following the change in absorbance at 348 nm of 4-NPA to 4-nitrophenylate ion over a period of 3 min at 25°C using a spect...More data for this Ligand-Target Pair
Affinity DataKi: 970nMpH: 7.4Assay Description:CA activity was assayed by following the change in absorbance at 348 nm of 4-NPA to 4-nitrophenylate ion over a period of 3 min at 25°C using a spect...More data for this Ligand-Target Pair
Affinity DataKi: 1.12E+3nMpH: 7.4Assay Description:CA activity was assayed by following the change in absorbance at 348 nm of 4-NPA to 4-nitrophenylate ion over a period of 3 min at 25°C using a spect...More data for this Ligand-Target Pair
Affinity DataKi: 1.49E+3nMpH: 7.4Assay Description:CA activity was assayed by following the change in absorbance at 348 nm of 4-NPA to 4-nitrophenylate ion over a period of 3 min at 25°C using a spect...More data for this Ligand-Target Pair
Affinity DataKi: 2.21E+3nMpH: 7.4Assay Description:CA activity was assayed by following the change in absorbance at 348 nm of 4-NPA to 4-nitrophenylate ion over a period of 3 min at 25°C using a spect...More data for this Ligand-Target Pair
Affinity DataKi: 1.48E+5nM IC50: 2.12E+5nMpH: 7.0Assay Description:Guinea pig AO activity was assayed spectrophotometrically using phenanthridine as a substrate at 322 nm. All spectrophotometric determinations were c...More data for this Ligand-Target Pair
Affinity DataIC50: >5.00E+4nMAssay Description:Inhibition of rat serum Butyrylcholine esterase using butyrylthiocholine as substrate incubated for 15 mins by DTNB reagent based Ellman's methodMore data for this Ligand-Target Pair
Affinity DataIC50: 1.35E+4nMpH: 7.6 T: 22°CAssay Description:Ligand binding was determined using a scintillation proximity assay with streptavidin-coated polyvinyltoluene scintillation beads (Amersham) and biot...More data for this Ligand-Target Pair
Affinity DataIC50: 7.23E+4nMpH: 7.6 T: 22°CAssay Description:Ligand binding was determined using a scintillation proximity assay with streptavidin-coated polyvinyltoluene scintillation beads (Amersham) and biot...More data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+3nMAssay Description:Inhibition of porcine DPP4 after 30 mins by luminescence assayMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Jishou University
Curated by ChEMBL
Jishou University
Curated by ChEMBL
Affinity DataIC50: 3.28E+5nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease-mediated ammonia production preincubated for 1.5 hrs by indophenol methodMore data for this Ligand-Target Pair
Affinity DataIC50: >5.00E+4nMAssay Description:Inhibition of rat cortex AChE using acetylthiocholine as substrate incubated for 15 mins by DTNB reagent based Ellman's methodMore data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 6 hits for monomerid = 19461
Found 6 hits for monomerid = 19461
CellTranscriptional Regulator TtgR Mutant R176G(Pseudomonas putida (g-Proteobacteria))
Imperial College
Imperial College
ITC DataΔG°: -6.29kcal/mole −TΔS°: 18.6kcal/mole ΔH°: -24.9kcal/mole logk: 3.50E+4
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -7.01kcal/mole −TΔS°: 2.48kcal/mole ΔH°: -9.49kcal/mole logk: 1.15E+5
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -5.80kcal/mole −TΔS°: -2.35kcal/mole ΔH°: -3.44kcal/mole logk: 1.54E+4
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -6.59kcal/mole −TΔS°: 4.19kcal/mole ΔH°: -10.8kcal/mole logk: 5.50E+4
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -6.96kcal/mole −TΔS°: 20.1kcal/mole ΔH°: -27.2kcal/mole logk: 1.07E+5
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -6.96kcal/mole −TΔS°: 7.22kcal/mole ΔH°: -14.2kcal/mole logk: 1.06E+5
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C