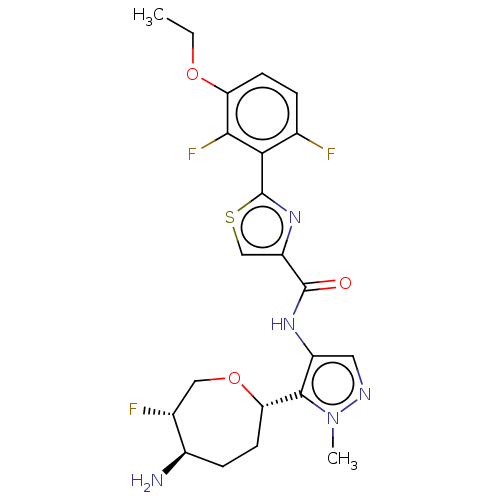
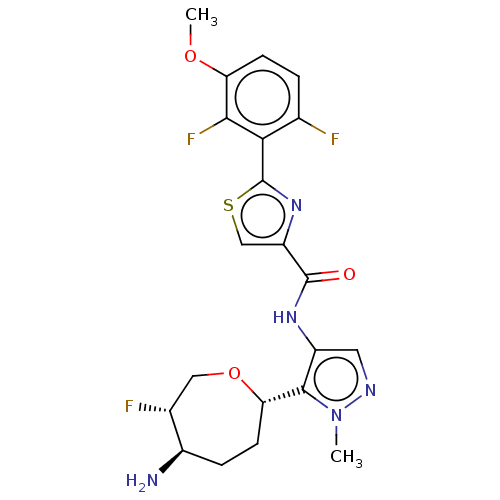
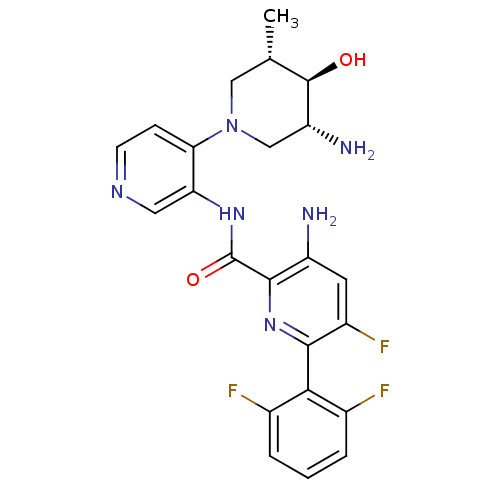
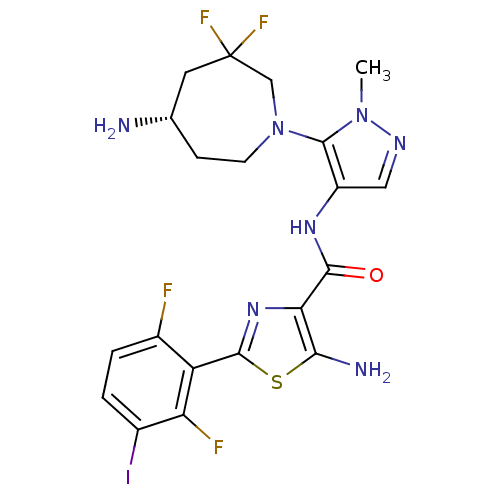
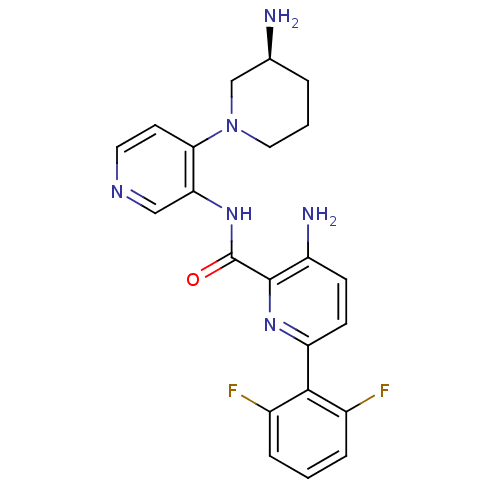
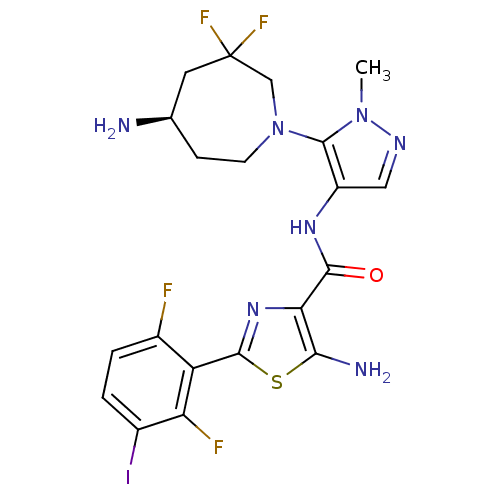
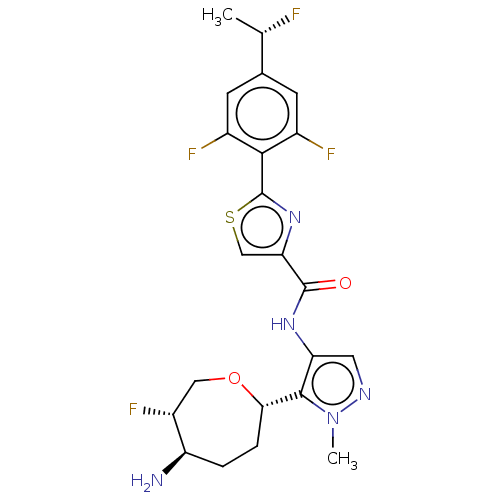
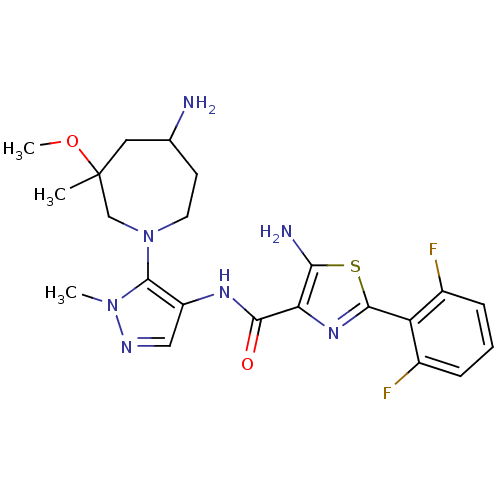
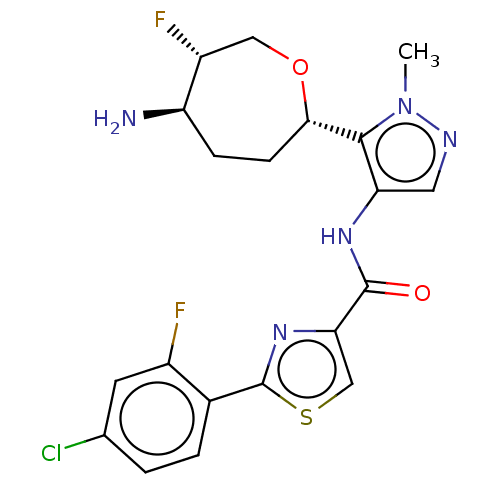
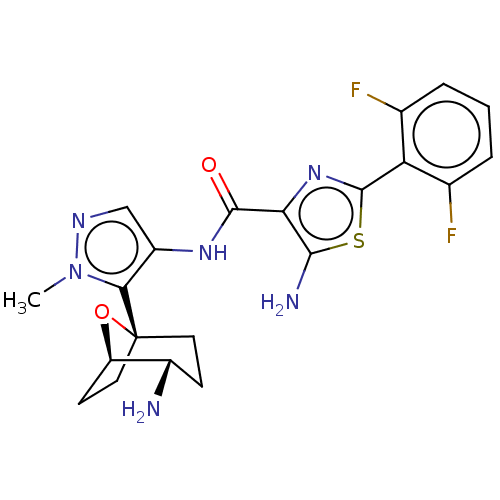
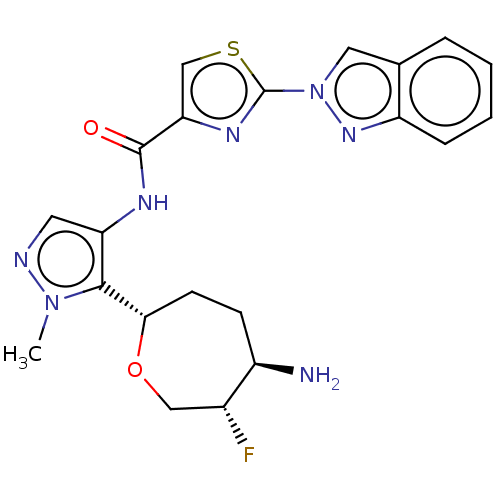
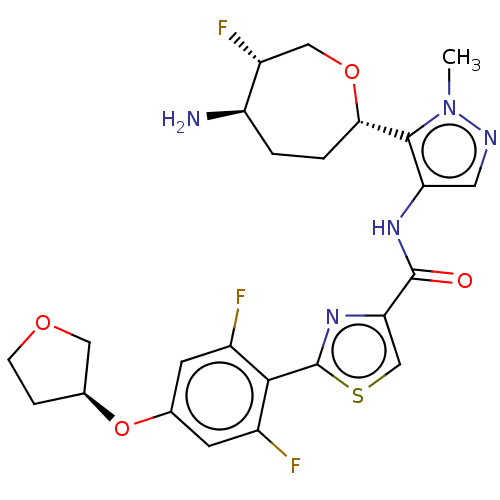
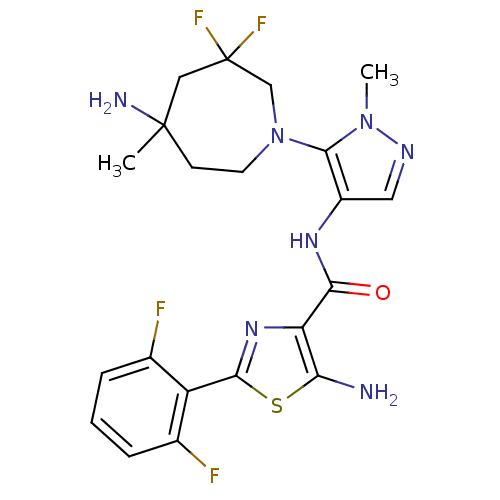
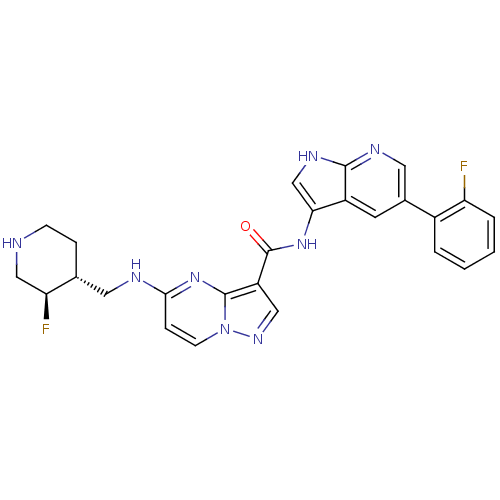
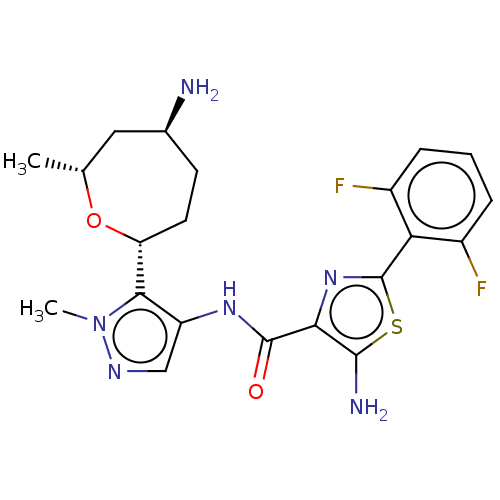
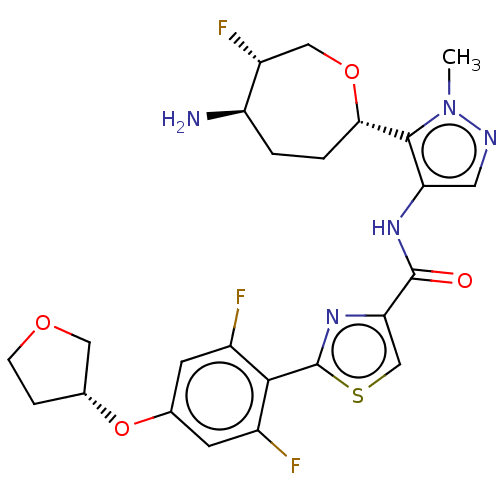
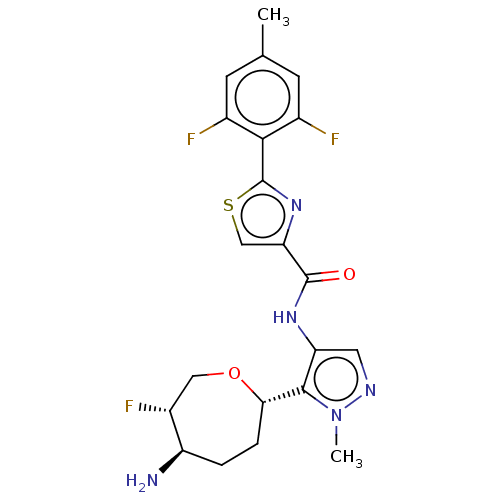
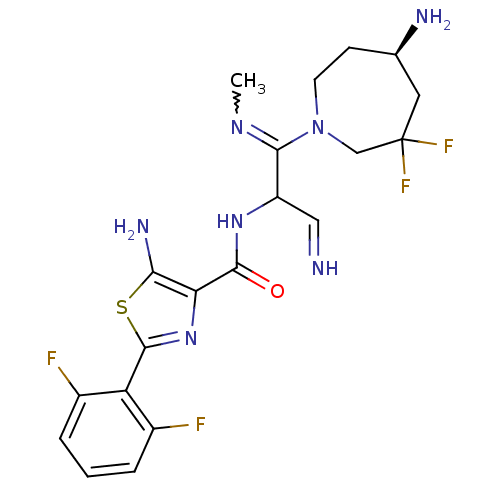
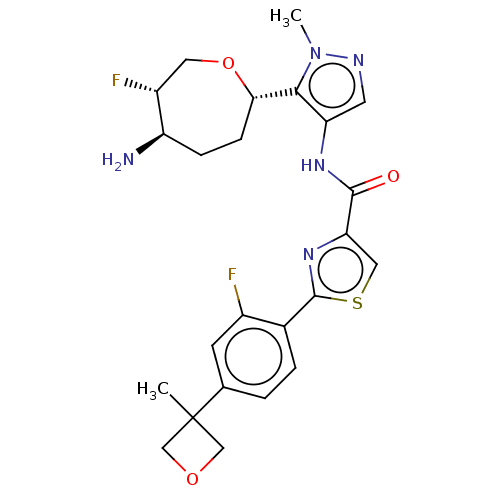
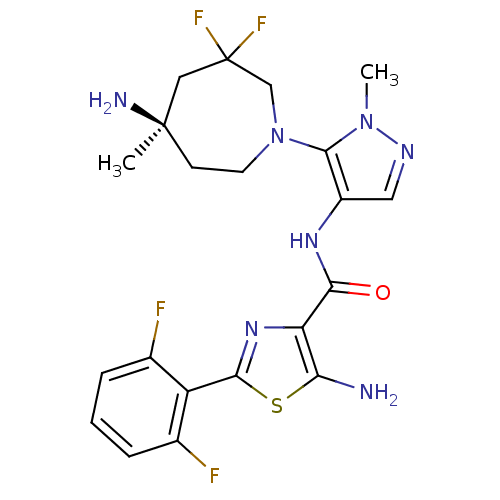
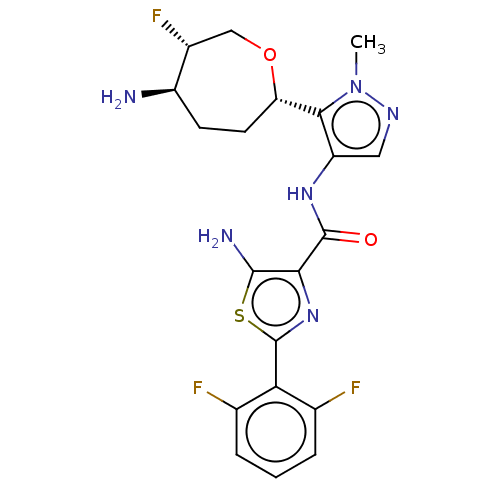
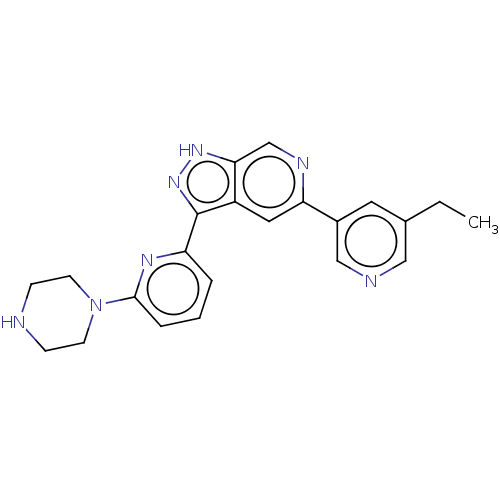
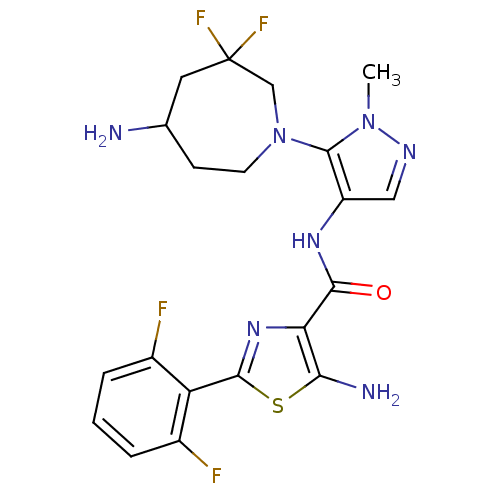
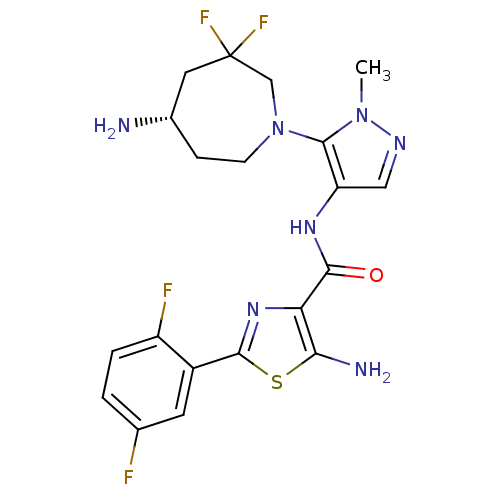
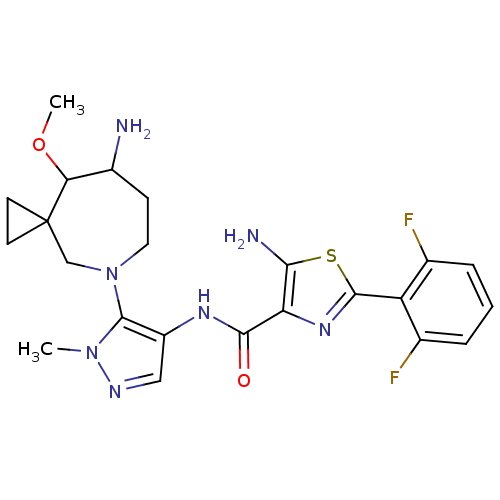
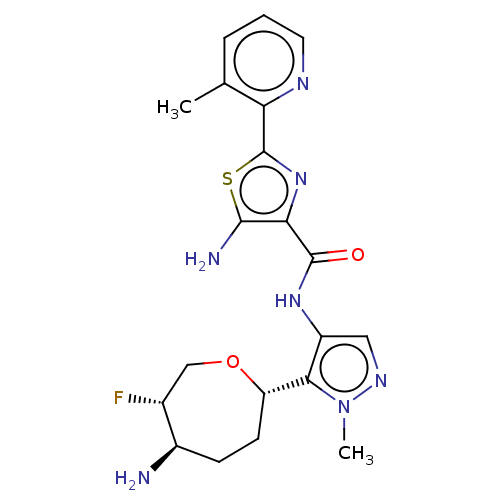
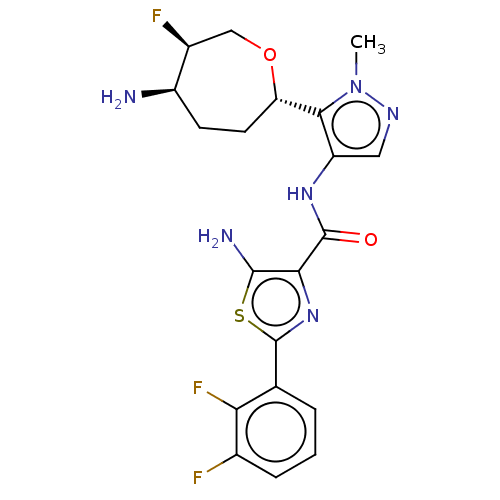
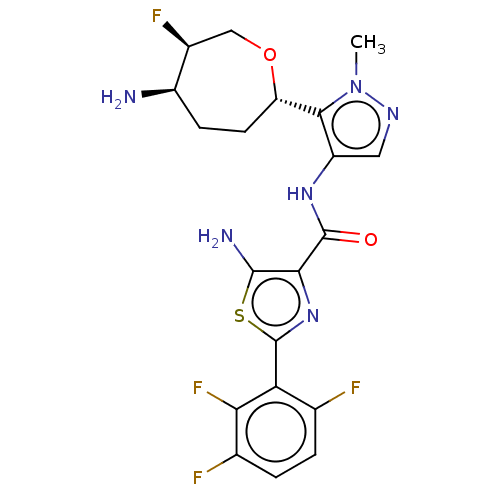
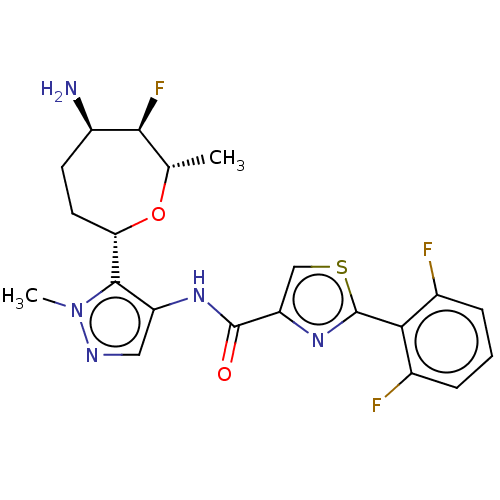
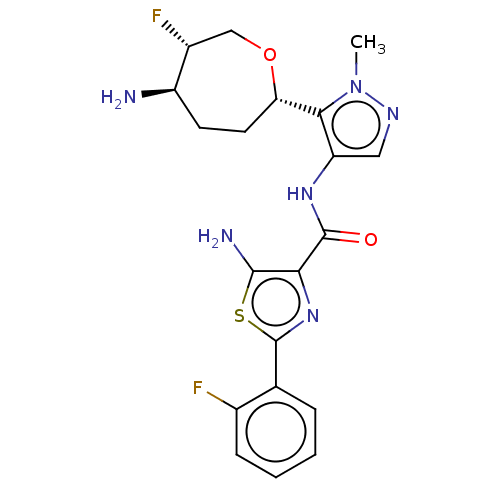
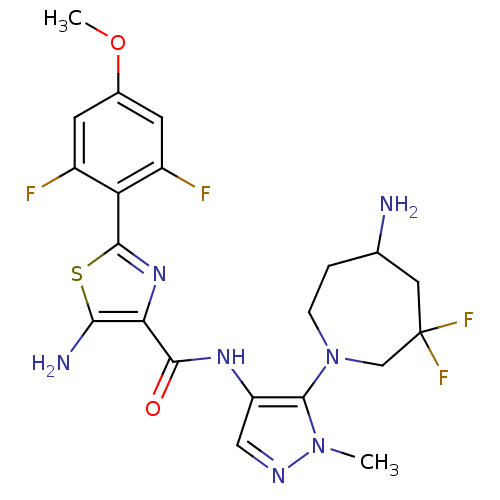
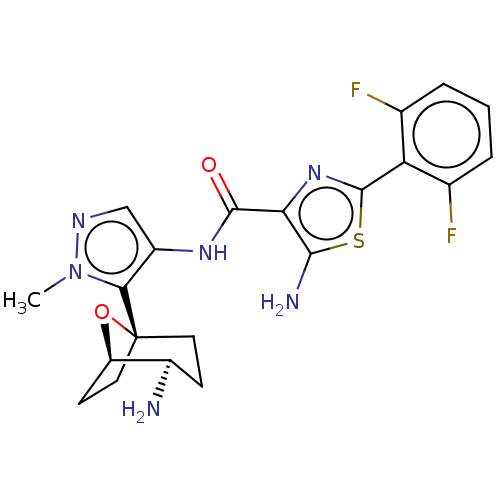
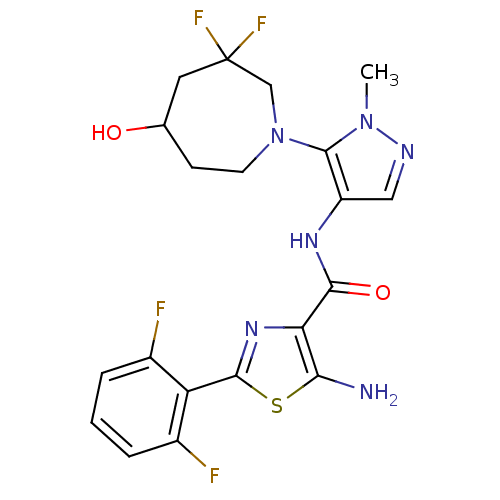
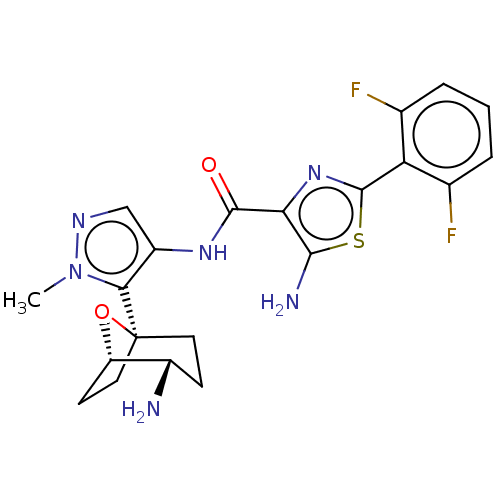
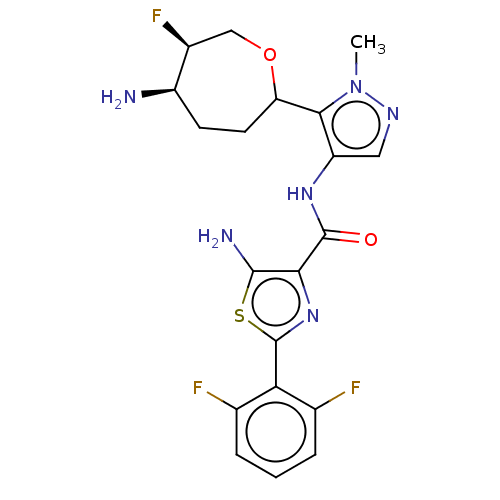
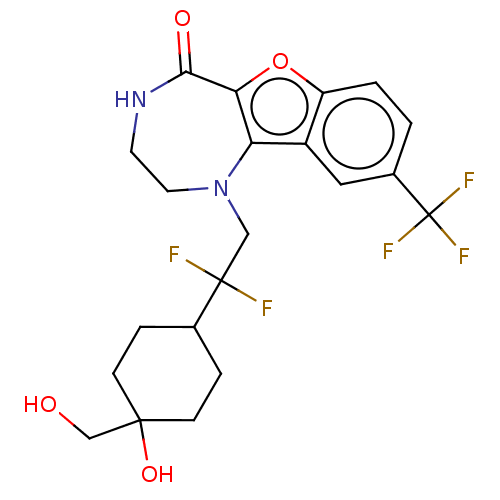
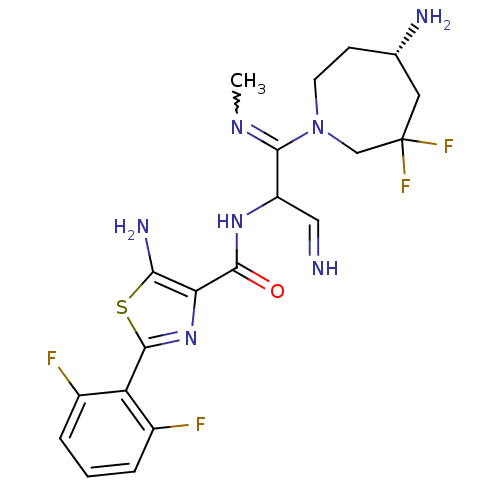
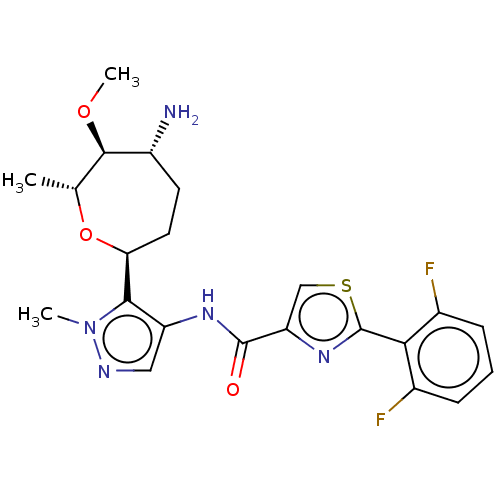
Report error Found 7639 Enz. Inhib. hit(s) with Target = 'Serine/threonine-protein kinase pim-1'
Affinity DataKi: 0.00100nM ΔG°: -68.5kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00100nM ΔG°: -68.5kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00100nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00100nM ΔG°: -68.5kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00100nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMAssay Description:Inhibition of PIM1 (unknown origin) using Biotin-AGAGRSRHSSYPAGT-OH as substrate after 2 hrs by AlphaScreen assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMAssay Description:Determination of the Pim kinase activity of a Formula I compound is possible by a number of direct and indirect detection methods. Certain exemplary ...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nM ΔG°: -66.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nM ΔG°: -66.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00200nM ΔG°: -66.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00224nM ΔG°: -66.5kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nMAssay Description:Inhibition of PIM1 (unknown origin) using FAM-pimtide as substrate after 90 mins by spectrophotometry in presence of ATPMore data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nM ΔG°: -65.8kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00300nMAssay Description:Inhibition of recombinant human PIM1 using FAM-pimtide as substrate after 90 mins by Z-LYTE assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.00342nM ΔG°: -65.4kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nMAssay Description:Determination of the Pim kinase activity of a Formula I compound is possible by a number of direct and indirect detection methods. Certain exemplary ...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nMpH: 7.2Assay Description:PIM-1, PIM-2, and PIM-3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J....More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nM ΔG°: -65.1kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nM ΔG°: -65.1kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00400nM ΔG°: -65.1kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00500nMAssay Description:Determination of the Pim kinase activity of a Formula I compound is possible by a number of direct and indirect detection methods. Certain exemplary ...More data for this Ligand-Target Pair
Affinity DataKi: 0.00500nMAssay Description:Inhibition of recombinant human PIM1 using FAM-pimtide as substrate after 90 mins by Z-LYTE assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.00509nM ΔG°: -64.5kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMAssay Description:Inhibition of recombinant human PIM1 using FAM-pimtide as substrate after 90 mins by Z-LYTE assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.00600nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nM ΔG°: -64.1kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataIC50: 0.00600nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nM ΔG°: -64.1kJ/molepH: 7.2 T: 2°CAssay Description:LC3K assay: PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Ch...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMAssay Description:The calculation of the Pim-1 inhibitory activity of the compound was performed using the following solution according to the protocol attached to ADP...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMAssay Description:Inhibition of PIM1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMAssay Description:Inhibition of PIM1 kinase (unknown origin) using Biotin-AGAGRSRHSSYPAGT-OH as substrate after 2 hrs by alphascreen assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMpH: 7.2Assay Description:PIM-1, -2, and -3 enzymes were generated as fusion proteins expressed in bacteria and purified by IMAC column chromatography (Sun, X., Chiu, J. F., a...More data for this Ligand-Target Pair
Affinity DataKi: 0.00600nMAssay Description:Determination of the Pim kinase activity of a Formula I compound is possible by a number of direct and indirect detection methods. Certain exemplary ...More data for this Ligand-Target Pair