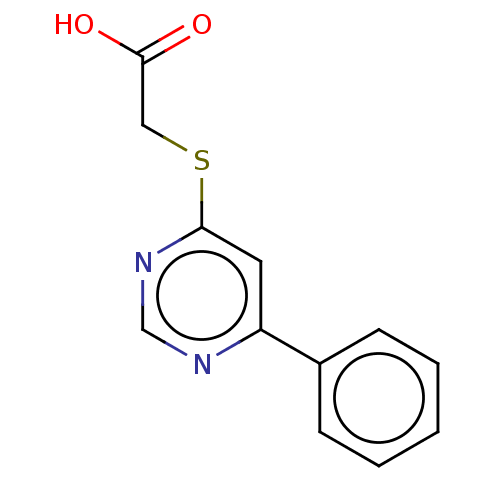
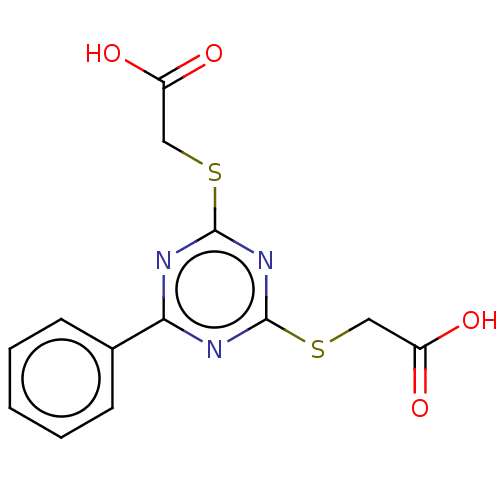
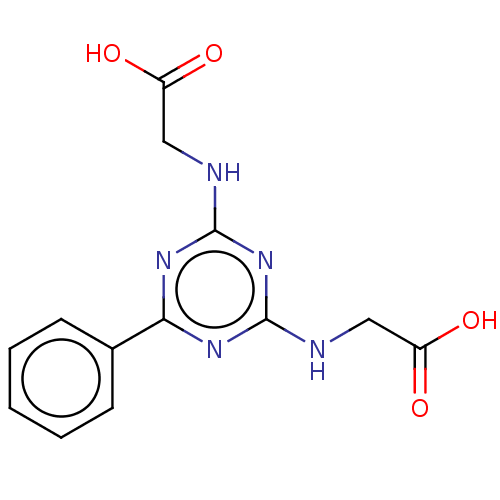
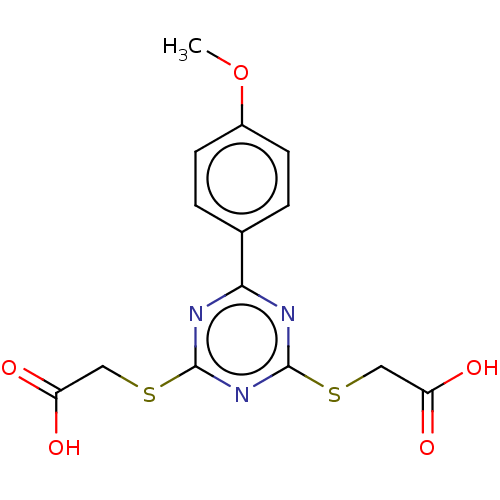
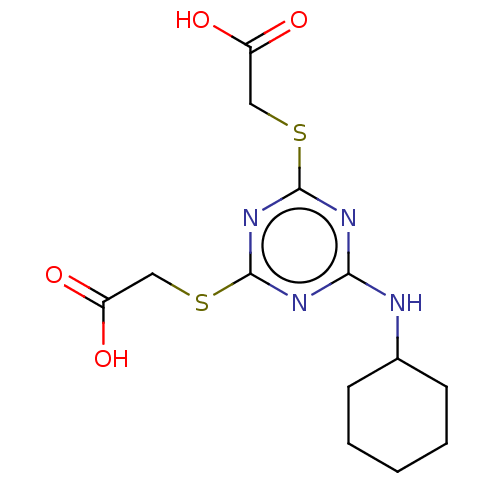
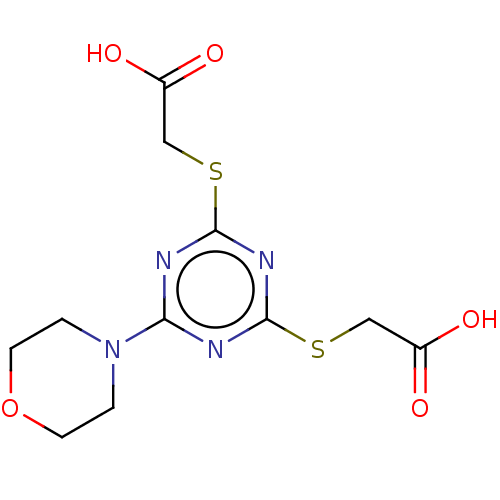
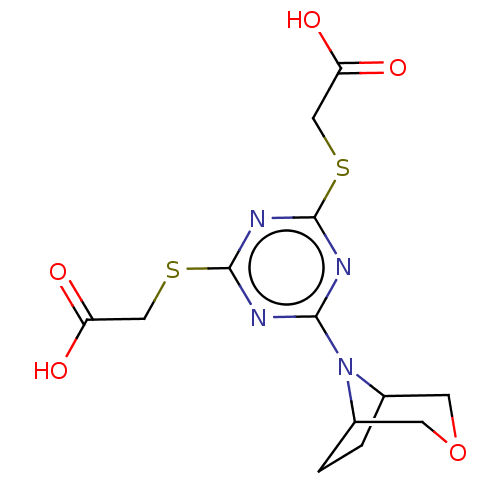
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.69E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 3.02E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 9.00E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.58E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 3.55E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.58E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.56E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 3.83E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 6.46E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 3.57E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 3.06E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.80E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.33E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.34E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.64E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 8.00E+4nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 2.51E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.36E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.70E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.03E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 2.34E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 2.33E+5nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair
TargetStromal cell-derived factor 1(Homo sapiens (Human))
Medical College Of Wisconsin
Curated by ChEMBL
Medical College Of Wisconsin
Curated by ChEMBL
Affinity DataKd: 1.20E+6nMAssay Description:Binding affinity to wild type CXCL12 (unknown origin) by NMR spectroscopyMore data for this Ligand-Target Pair