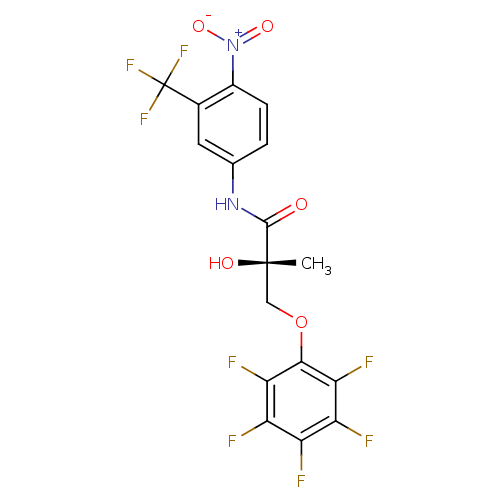
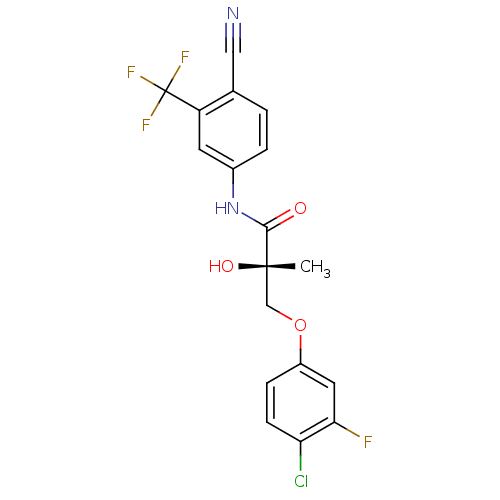
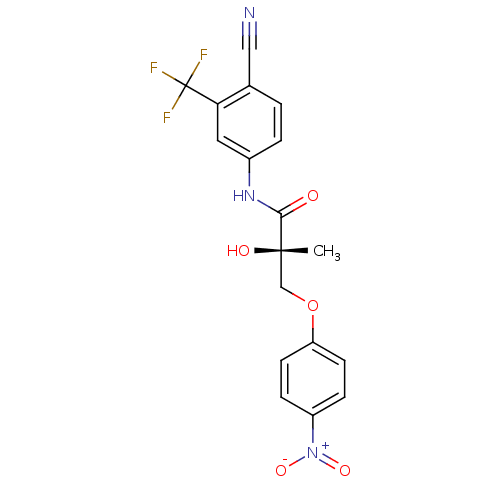
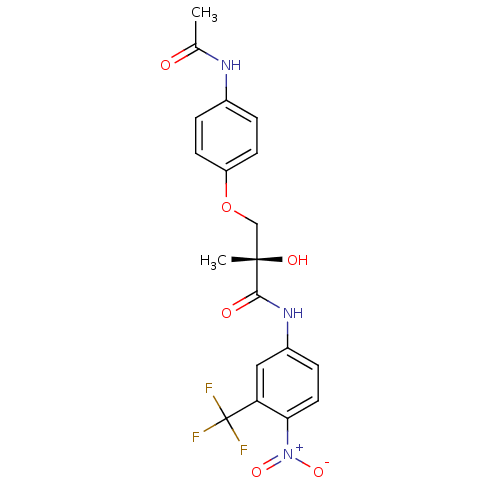
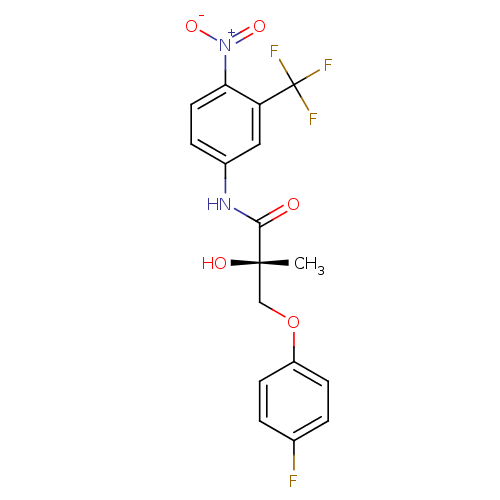
Affinity DataKi: 0.270nM ΔG°: -50.8kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
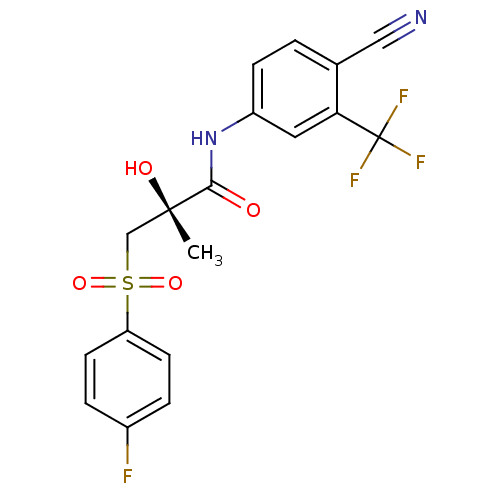
Affinity DataKi: 0.540nM ΔG°: -49.2kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
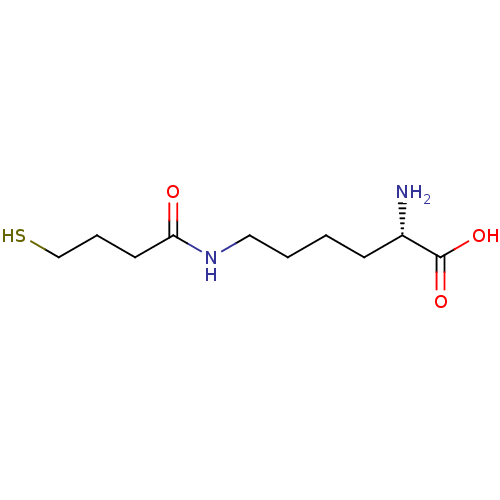
Affinity DataKi: 1.40nM ΔG°: -47.0kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 1.70nM ΔG°: -46.5kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 2.5nM ΔG°: -45.6kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 4nM ΔG°: -44.6kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 6.10nM ΔG°: -43.6kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 11nM ΔG°: -42.2kJ/molepH: 7.4 T: 2°CAssay Description:The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (...More data for this Ligand-Target Pair
Affinity DataKi: 370nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 430nMAssay Description:Inhibition of ferrous substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 720nMAssay Description:Inhibition of ferrous substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 720nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Escherichia coli (strain K12))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 3.20E+3nMAssay Description:Inhibition of cobalt substituted Escherichia coli LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Vibrio harveyi (strain ATCC BAA-1116 / BB120))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 9.70E+3nMAssay Description:Inhibition of cobalt substituted Vibrio harveyi LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.06E+4nMAssay Description:Inhibition of zinc substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Escherichia coli (strain K12))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.27E+4nMAssay Description:Inhibition of cobalt substituted Escherichia coli LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Vibrio harveyi (strain ATCC BAA-1116 / BB120))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 1.28E+4nMAssay Description:Inhibition of cobalt substituted Vibrio harveyi LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.96E+4nMAssay Description:Inhibition of zinc substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 6.10E+4nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 6.80E+4nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 7.00E+4nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.32E+5nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.47E+5nMAssay Description:Inhibition of ferrous substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.55E+5nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 1.56E+5nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 4.73E+5nMAssay Description:Inhibition of cobalt substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Vibrio harveyi (strain ATCC BAA-1116 / BB120))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 5.50E+5nMAssay Description:Inhibition of cobalt substituted Vibrio harveyi LuxSMore data for this Ligand-Target Pair
TargetS-ribosylhomocysteine lyase(Escherichia coli (strain K12))
The Ohio State University
Curated by ChEMBL
The Ohio State University
Curated by ChEMBL
Affinity DataKi: 7.20E+5nMAssay Description:Inhibition of cobalt substituted Escherichia coli LuxSMore data for this Ligand-Target Pair
Affinity DataKi: 2.40E+6nMAssay Description:Inhibition of zinc substituted Bacillus subtilis LuxSMore data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)