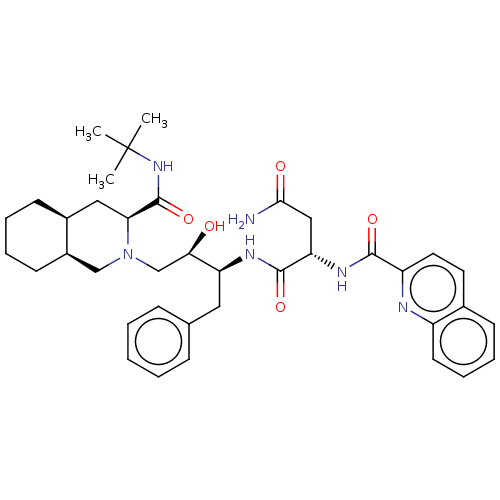
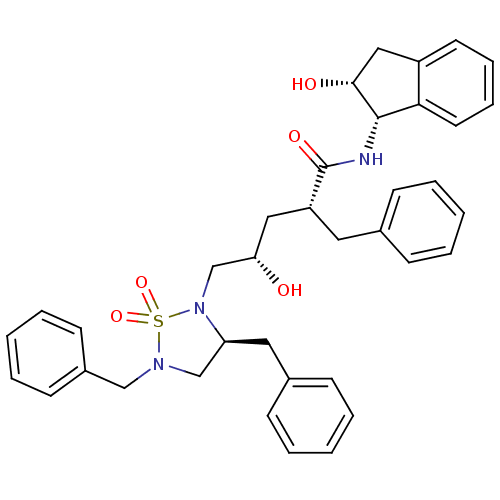
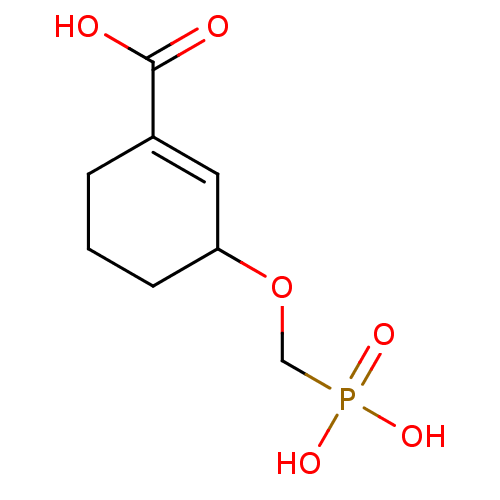
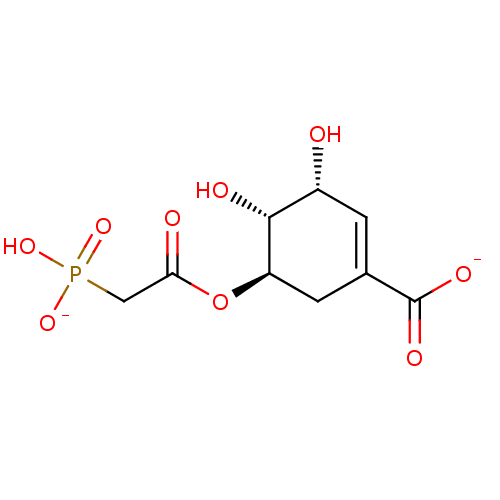
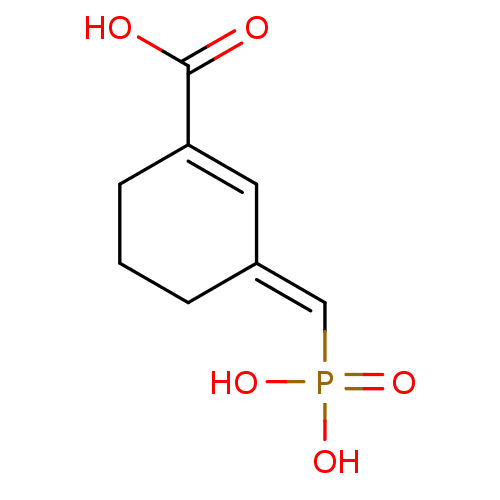
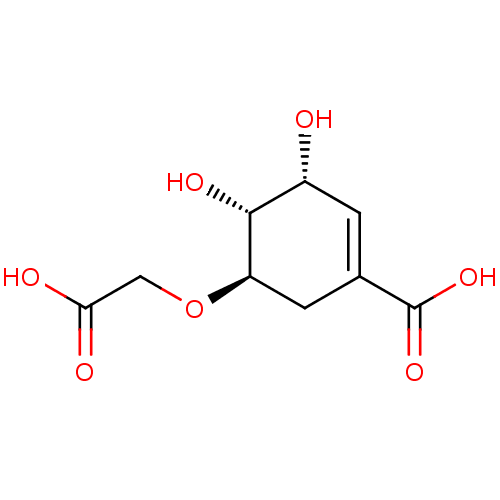
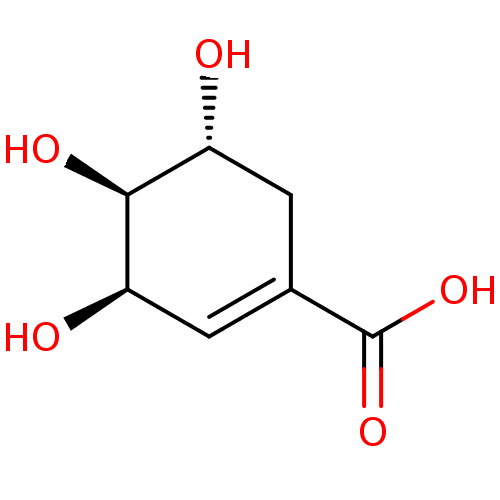
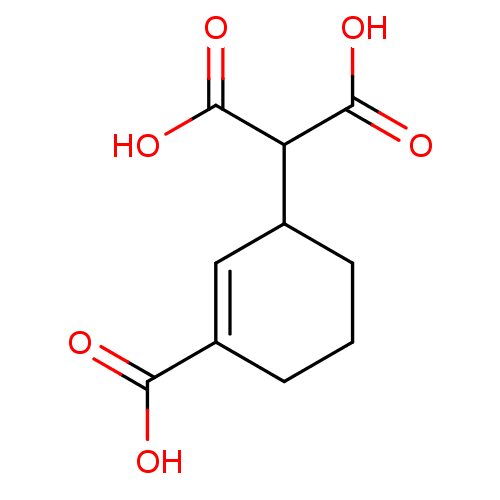
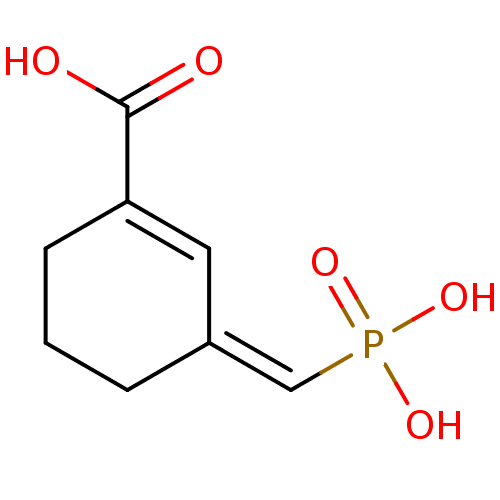
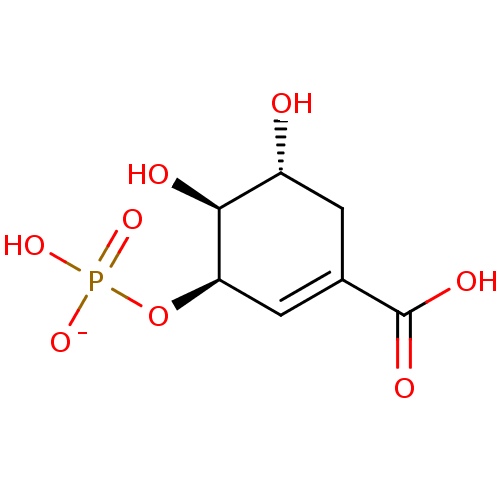
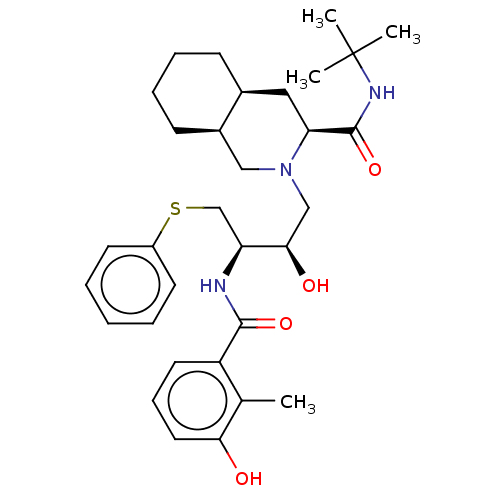
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0100nM ΔG°: -62.8kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
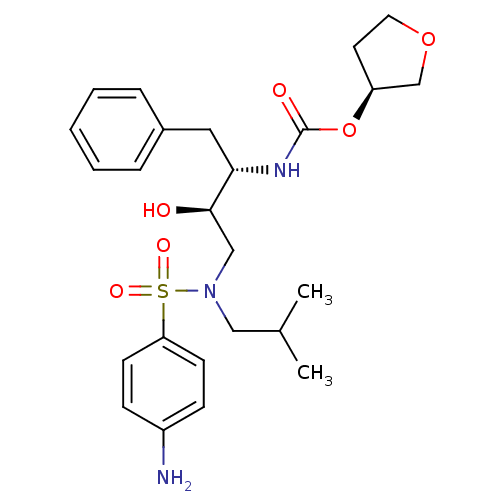
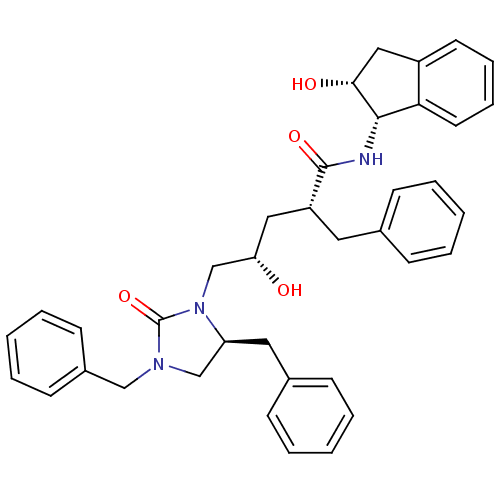
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0200nM ΔG°: -61.1kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
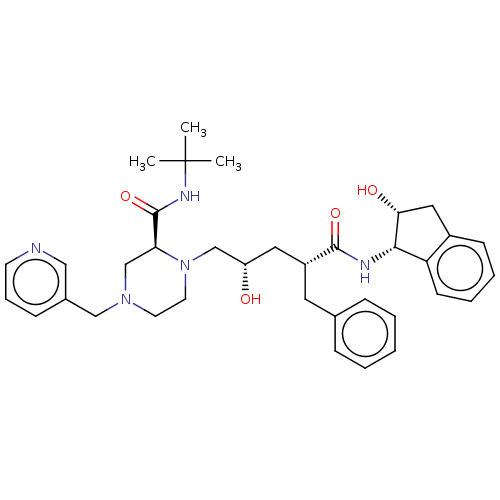
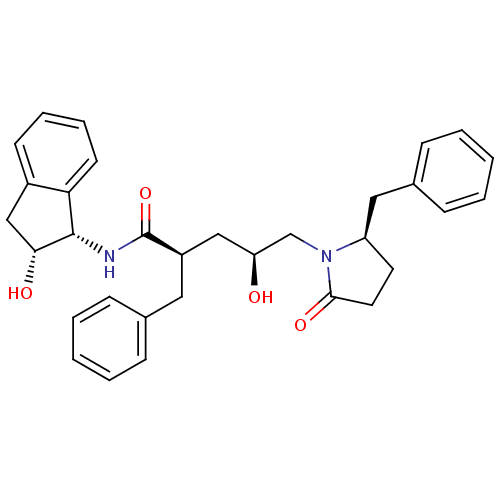
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0400nM ΔG°: -59.3kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
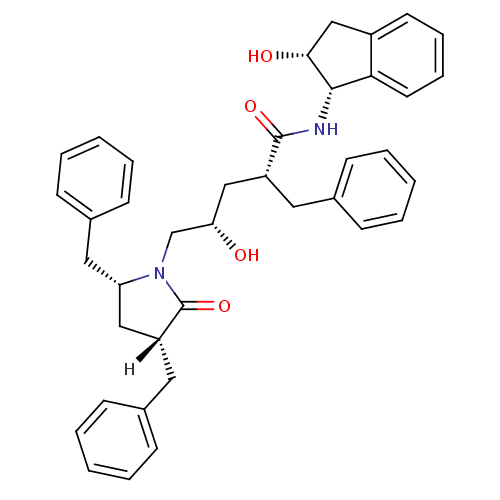
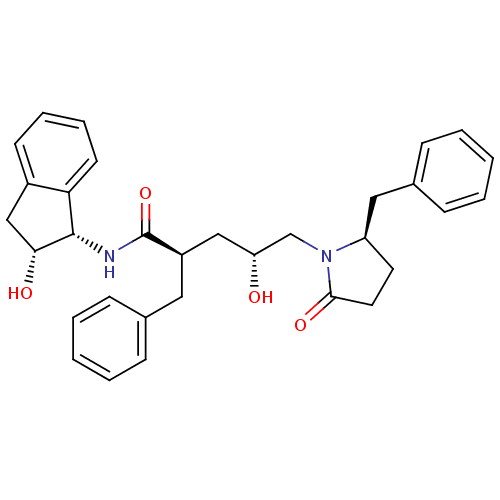
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0400nM ΔG°: -59.3kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0500nM ΔG°: -58.8kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.0700nM ΔG°: -58.0kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: >0.5nM ΔG°: >-53.1kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 0.800nM ΔG°: -51.9kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 1.20nM ΔG°: -50.9kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 16nM ΔG°: -44.5kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 330nM ΔG°: -37.0kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
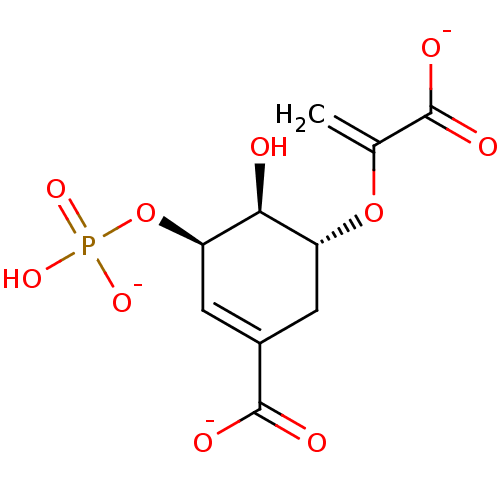
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataKi: 1.50E+3nMAssay Description:Dissociation constant of the compound was calculated for E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Glaxosmithkline
Glaxosmithkline
Affinity DataKi: 4.80E+3nM ΔG°: -30.4kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
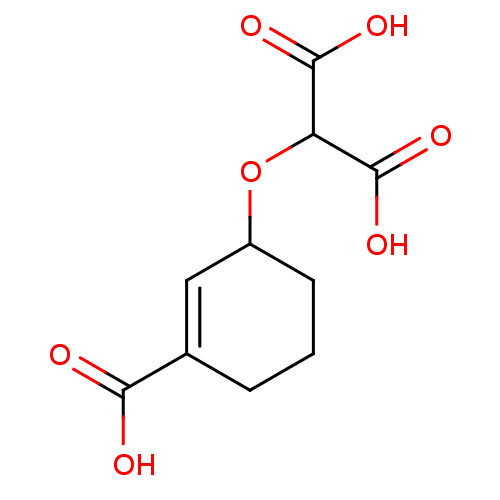
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: 1.80E+5nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: 7.50E+6nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
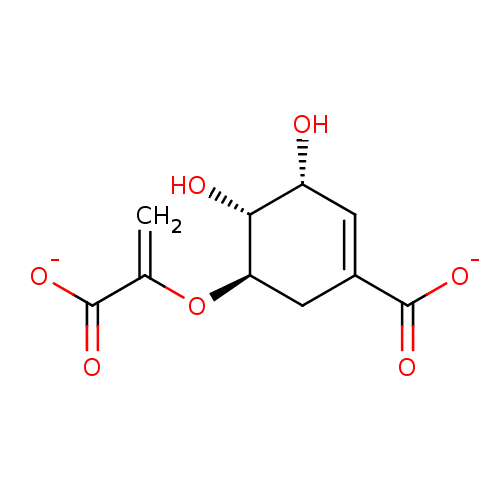
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: 1.50E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
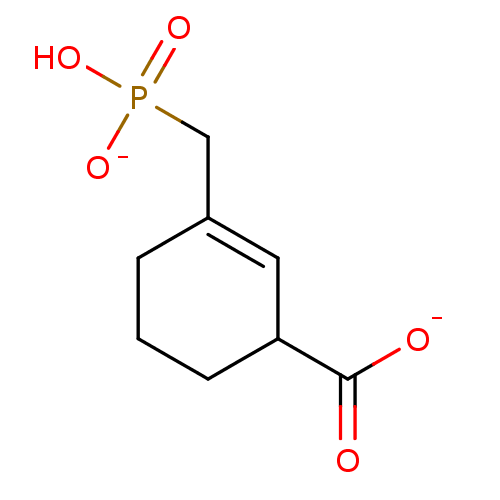
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataIC50: >9.00E+7nMAssay Description:Inhibition of E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataKd: 1.00E+3nMAssay Description:Binding affinity of the compound was determined against E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) synthaseMore data for this Ligand-Target Pair
Target3-phosphoshikimate 1-carboxyvinyltransferase(Escherichia coli (strain K12))
TBA
Curated by ChEMBL
TBA
Curated by ChEMBL
Affinity DataKd: 7.00E+3nMAssay Description:Inhibitory activity of the compound was evaluated against E. coli EPSP(5-enolpyruvyl-shikimate-3-phosphate) SynthaseMore data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)