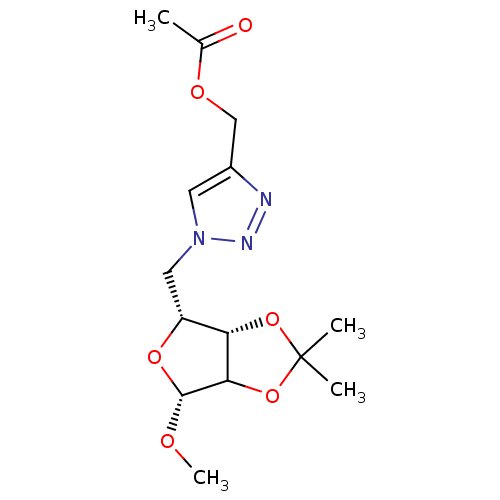
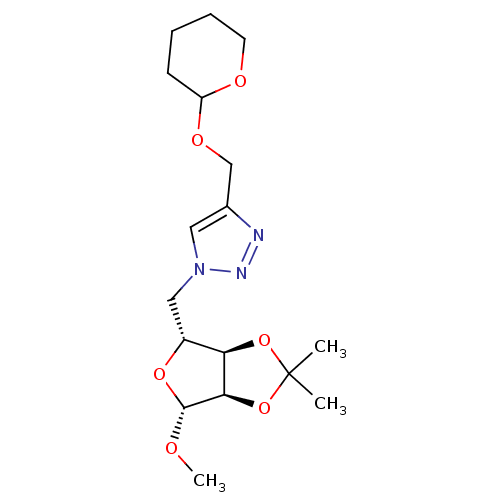
Affinity DataKi: 3.36E+4nM ΔG°: -26.6kJ/mole IC50: 3.58E+4nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
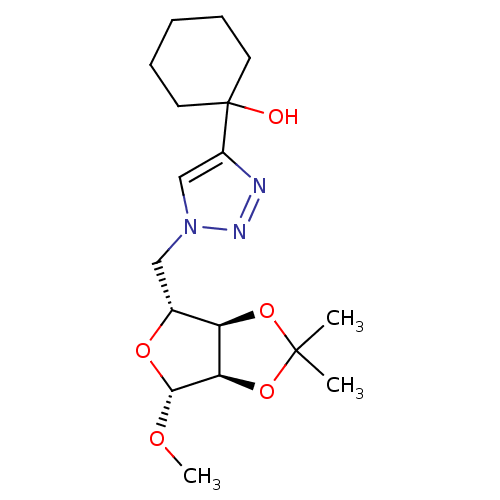
Affinity DataKi: 2.04E+5nM ΔG°: -21.9kJ/mole IC50: 8.89E+4nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
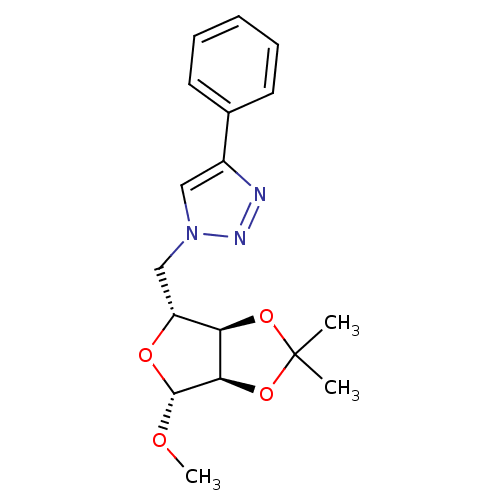
Affinity DataKi: 2.34E+5nM ΔG°: -21.6kJ/mole IC50: 1.25E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
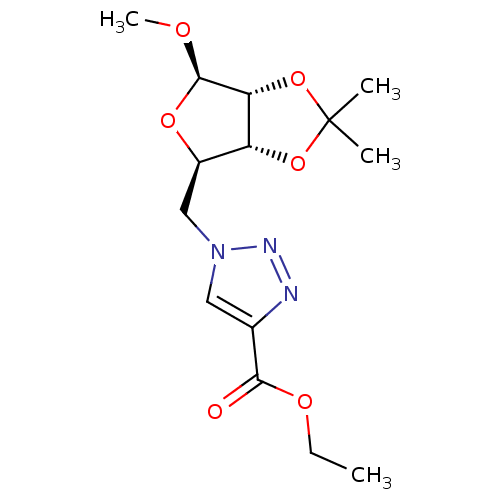
Affinity DataKi: 3.42E+5nM ΔG°: -20.6kJ/mole IC50: 1.50E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 7.80E+5nM ΔG°: -18.5kJ/mole IC50: 3.61E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
Affinity DataKi: 1.01E+6nM ΔG°: -17.8kJ/mole IC50: 3.56E+5nMpH: 7.0 T: 2°CAssay Description:To test whether GCTs share a common binding pocket on PPA with acarbose, we performed an experiment according to the method of Yonetani and Theorell.More data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
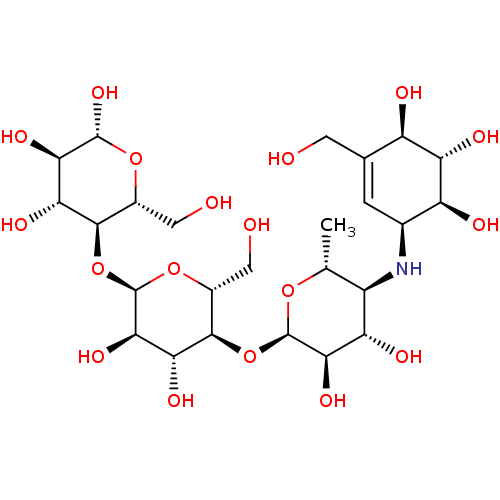
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 5.20E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 5.70E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 1.49E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 1.51E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 2.47E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
TargetAlpha-glucosidase MAL12(Saccharomyces cerevisiae)
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Universidade Federal Do Rio De Janeiro
Curated by ChEMBL
Affinity DataIC50: 1.09E+5nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair