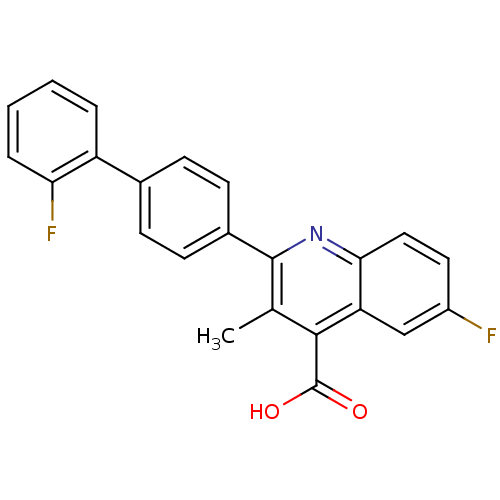
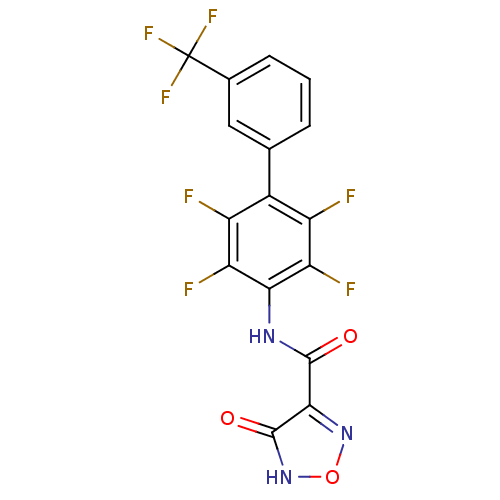
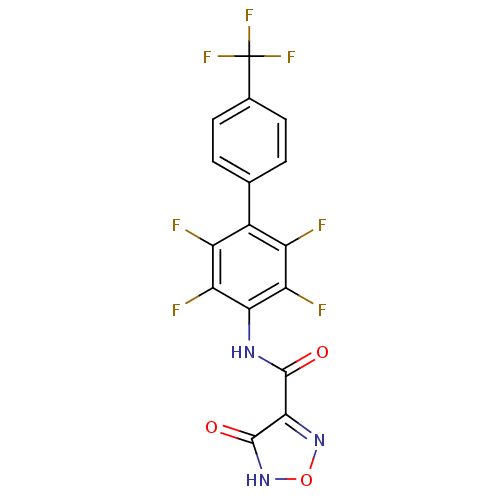
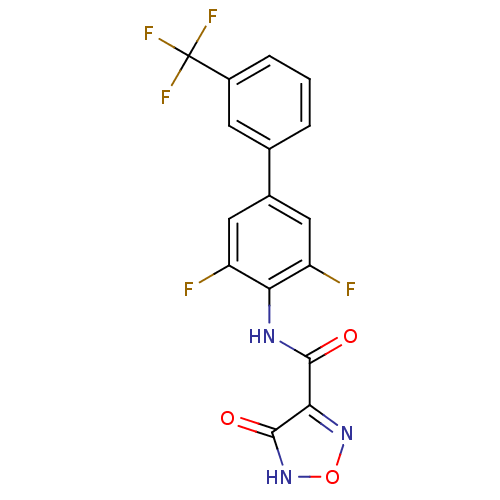
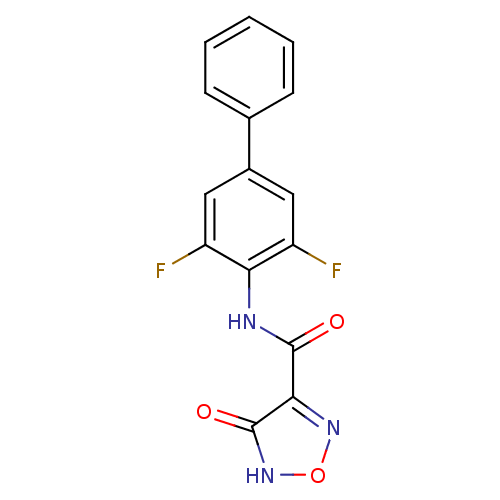
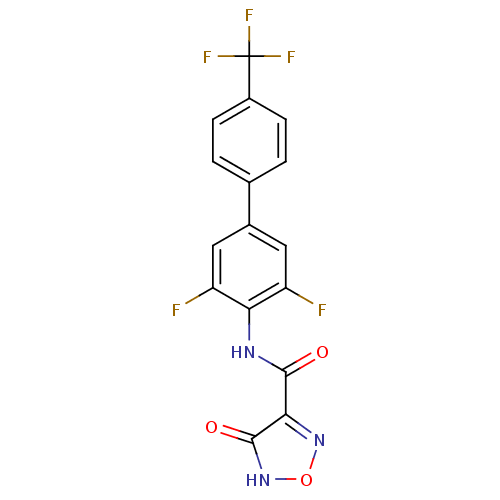
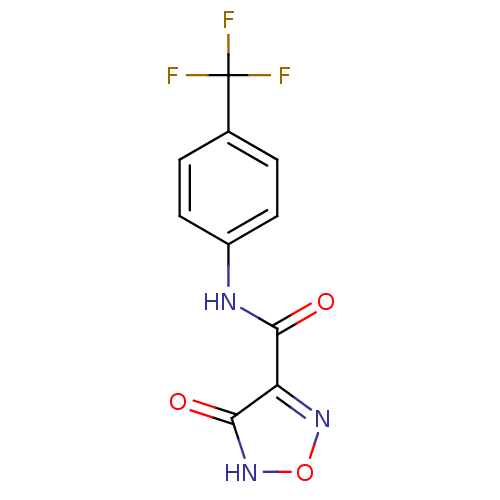
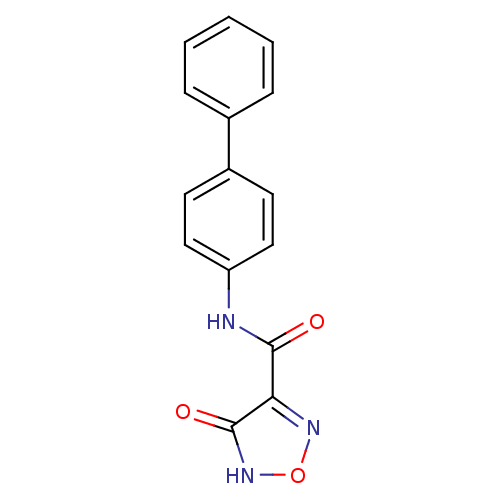
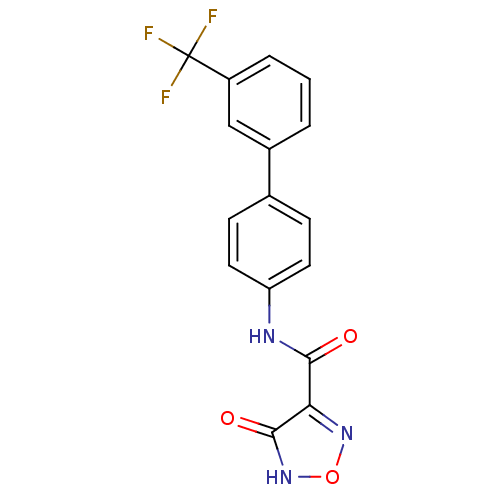
Affinity DataKi: 43nMAssay Description:Displacement of [3H]AMPA from rat iGluR2 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 43nMAssay Description:Displacement of [3H]AMPA from rat iGluR2 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 140nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR5 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 280nMAssay Description:Displacement of [3H]AMPA from rat iGluR2 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 330nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR6 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 490nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR7 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 8.10E+3nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR5 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 8.10E+3nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR5 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 2.00E+4nMAssay Description:Displacement of [3H]AMPA from rat iGluR2 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 4.00E+4nMAssay Description:Displacement of [3H]AMPA from rat iGluR2 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 4.10E+4nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR7 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 5.30E+4nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR5 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 6.70E+4nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR5 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR7 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR7 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR6 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR6 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+6nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR7 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+6nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR6 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+6nMAssay Description:Displacement of [3H]SYM2081 from rat iGluR6 receptor expressed in Sf9 cells baculovirus system after 1 to 2 hrs by liquid scintillation countingMore data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 20nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 33nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 50nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 66nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 120nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 490nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 870nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 880nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 2.30E+3nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 4.30E+3nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 1.10E+4nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 2.50E+4nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Biomedical Research Foundation Academy Of Athens (Brfaa)
Curated by ChEMBL
Biomedical Research Foundation Academy Of Athens (Brfaa)
Curated by ChEMBL
Affinity DataIC50: 2.60E+4nMAssay Description:Inhibition of PI3Kalpha catalytic domain (unknown origin) using PIP2 liposomes as substrate in presence of biotin-labeled PIP3 and GST-GRP1-PH by str...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 4.50E+4nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
TargetDihydroorotate dehydrogenase (quinone), mitochondrial(Rattus norvegicus (rat))
Universit£
Curated by ChEMBL
Universit£
Curated by ChEMBL
Affinity DataIC50: 4.90E+4nMAssay Description:Inhibition of dihydroorotate dehydrogenase in Wistar rat liver mitochondrial/microsomal membranes measured for 5 mins by 2,6-dichlorophenolindophenol...More data for this Ligand-Target Pair
Affinity DataIC50: 2.45E+5nMAssay Description:Antagonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as inhibition of (S)-Glu-induced intracellular calcium levels by FLIPR...More data for this Ligand-Target Pair
Affinity DataIC50: 2.45E+5nMAssay Description:Antagonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as inhibition of (S)-Glu-induced intracellular calcium levels by FLIPR...More data for this Ligand-Target Pair
Affinity DataIC50: 1.02E+6nMAssay Description:Antagonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as inhibition of (S)-Glu-induced intracellular calcium levels by FLIPR...More data for this Ligand-Target Pair
Affinity DataIC50: 1.02E+6nMAssay Description:Antagonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as inhibition of (S)-Glu-induced intracellular calcium levels by FLIPR...More data for this Ligand-Target Pair
Affinity DataEC50: 4.70E+4nMAssay Description:Agonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as change in intracellular calcium levels by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataEC50: 1.80E+3nMAssay Description:Agonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as change in intracellular calcium levels by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataEC50: 4.91E+5nMAssay Description:Activity at recombinant iGluR2 receptor expressed in Xenopus laevis oocytes using holding potential of -15 to -20 mV by TEVC electrophysiologyMore data for this Ligand-Target Pair
Affinity DataEC50: 4.89E+5nMAssay Description:Activity at recombinant iGluR2 receptor expressed in Xenopus laevis oocytes using holding potential of -15 to -20 mV by TEVC electrophysiologyMore data for this Ligand-Target Pair
Affinity DataEC50: 4.30E+4nMAssay Description:Activity at recombinant iGluR2 receptor expressed in Xenopus laevis oocytes using holding potential of -15 to -20 mV by TEVC electrophysiologyMore data for this Ligand-Target Pair
Affinity DataEC50: 4.30E+4nMAssay Description:Activity at recombinant iGluR2 receptor expressed in Xenopus laevis oocytes using holding potential of -15 to -20 mV by TEVC electrophysiologyMore data for this Ligand-Target Pair
Affinity DataEC50: 1.40E+4nMAssay Description:Activity at recombinant iGluR2 receptor expressed in Xenopus laevis oocytes using holding potential of -15 to -20 mV by TEVC electrophysiologyMore data for this Ligand-Target Pair
Affinity DataEC50: 4.70E+4nMAssay Description:Agonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as change in intracellular calcium levels by FLIPR assayMore data for this Ligand-Target Pair


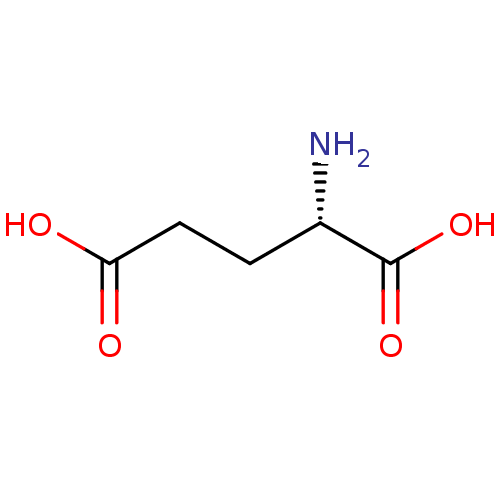
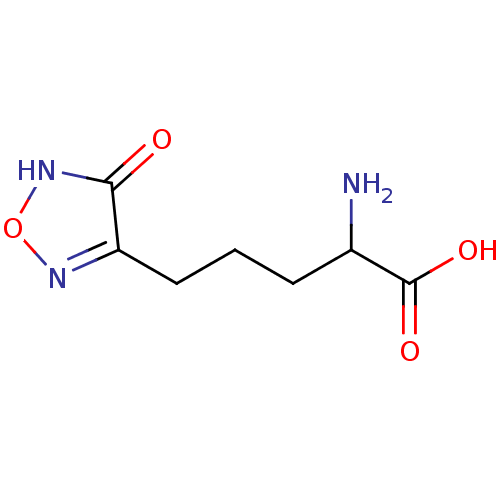
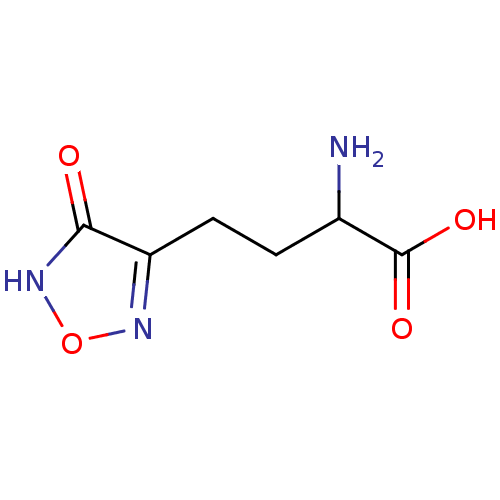
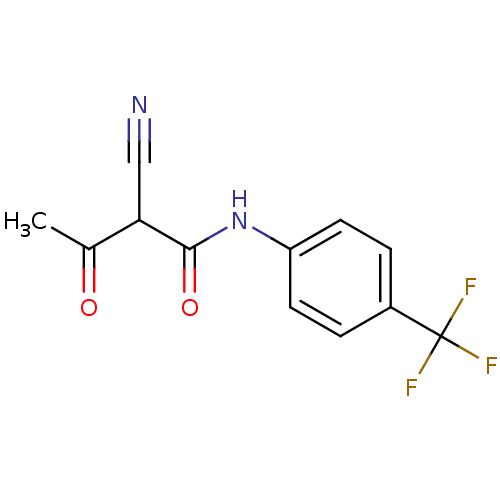
 3D Structure (crystal)
3D Structure (crystal)