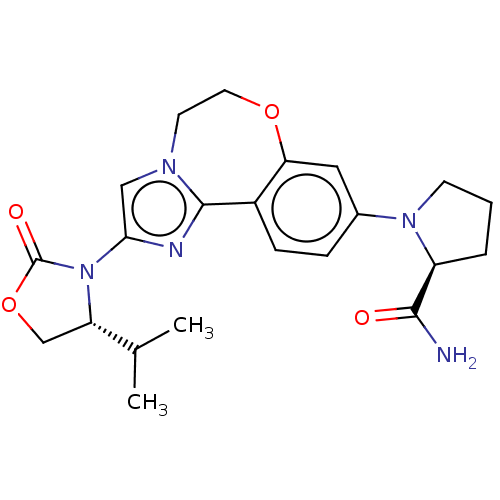
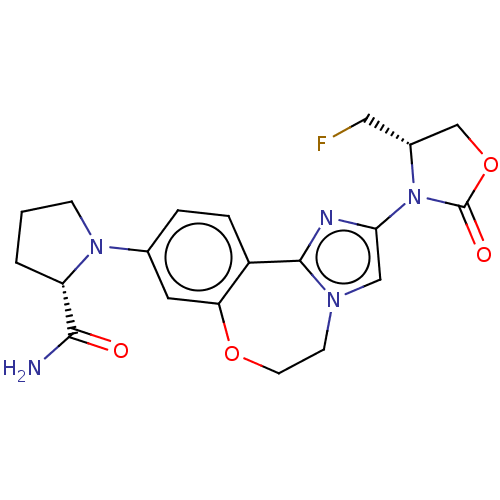
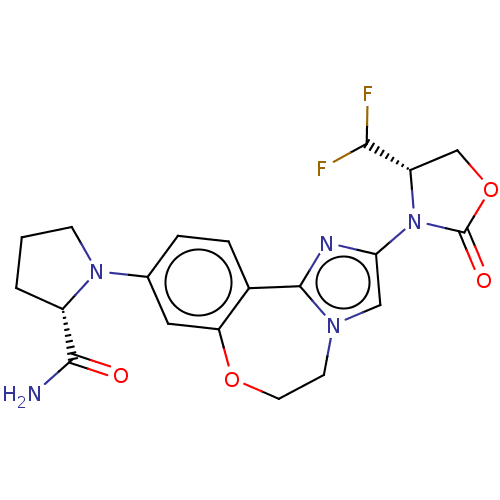
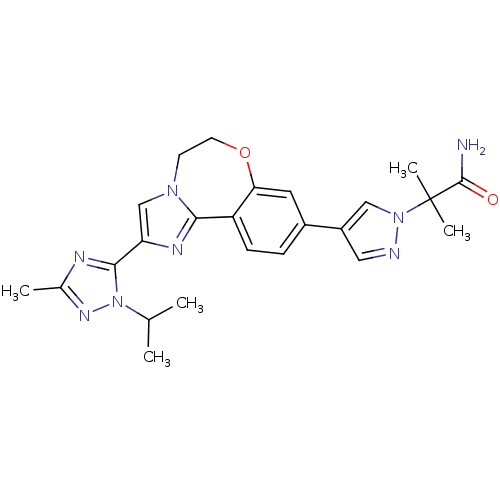
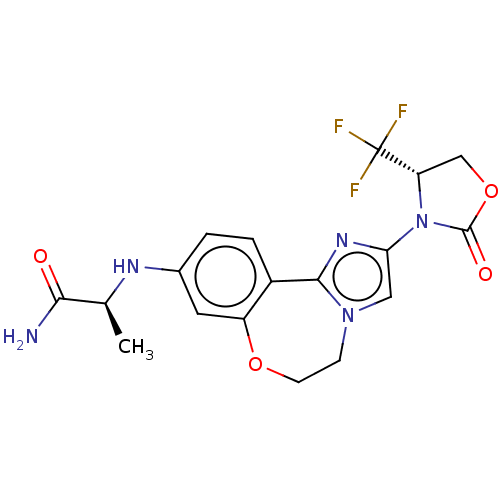
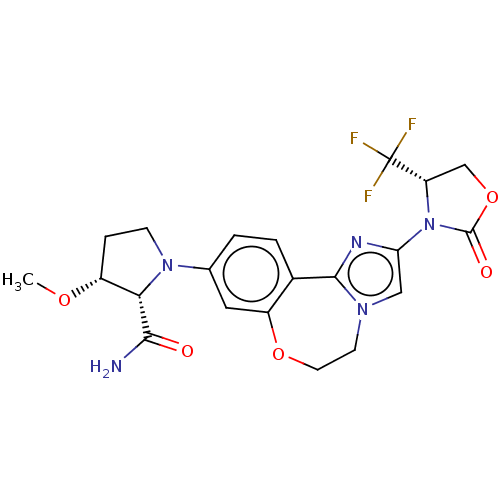
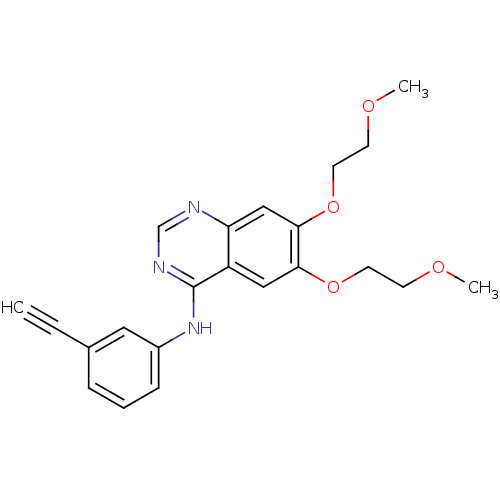
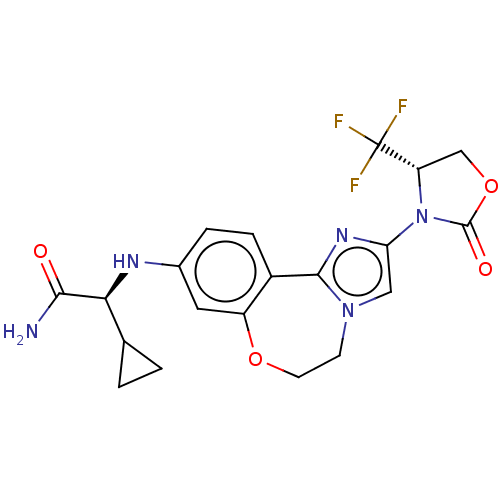
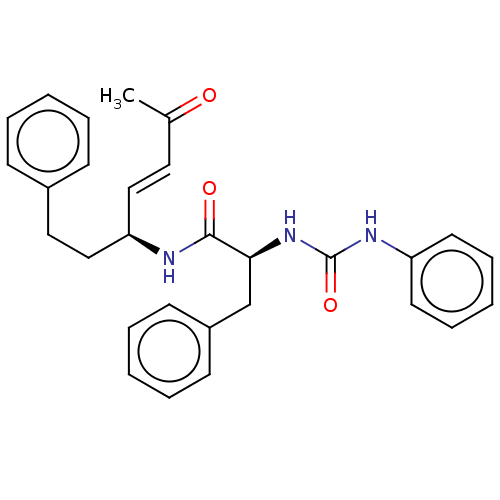
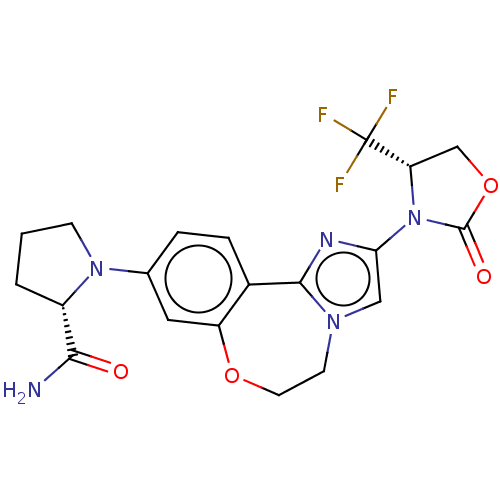
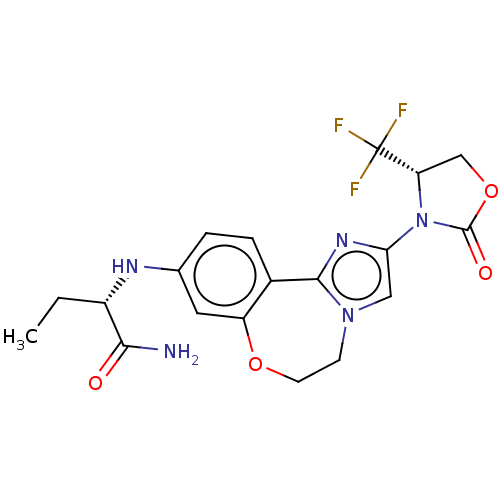
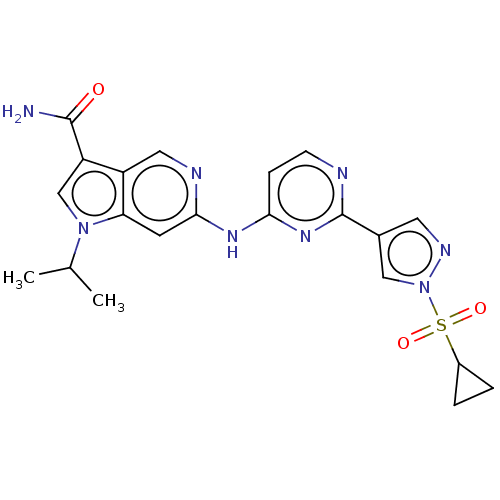
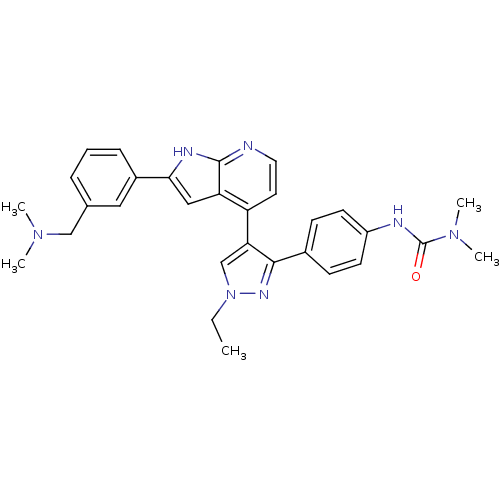
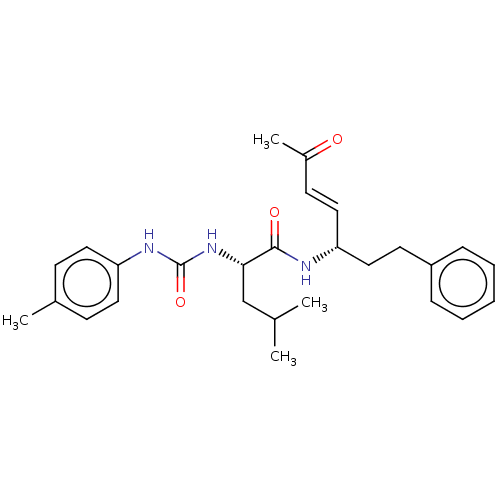
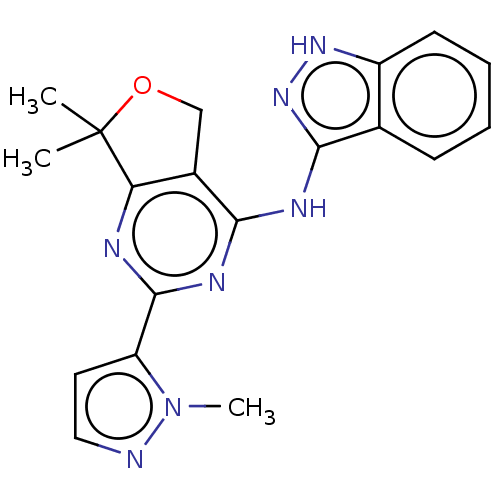
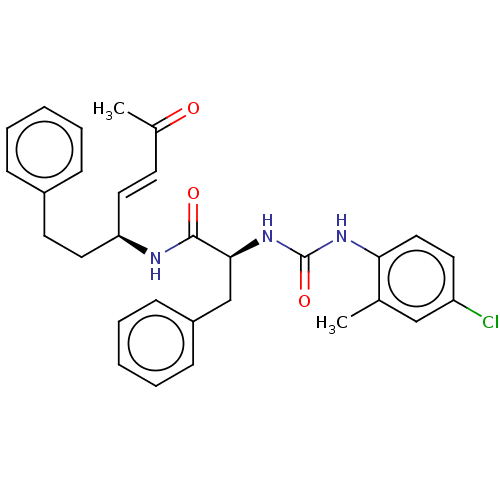
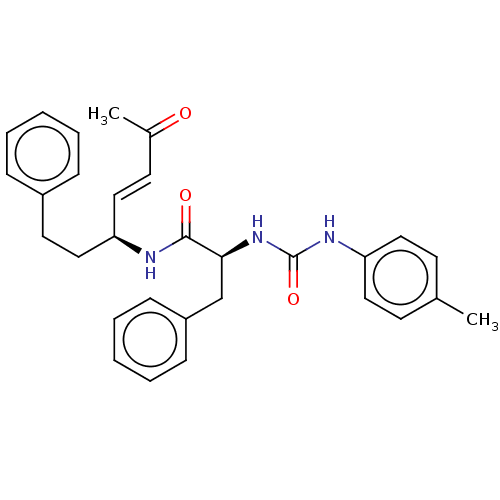
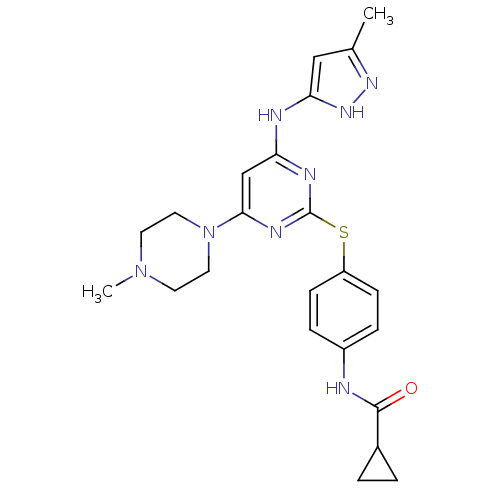
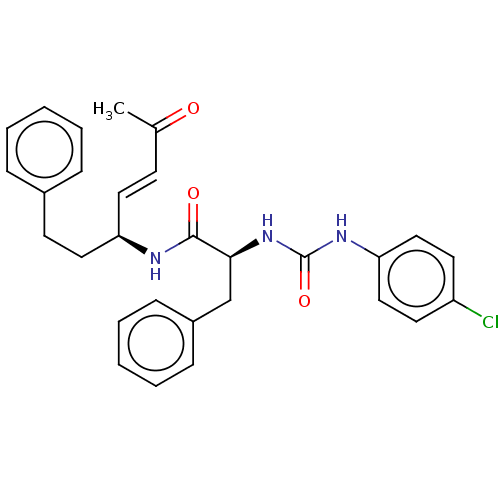
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0260nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0340nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0420nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0430nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0510nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0530nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0600nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0620nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0900nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0950nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.0970nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of EGFR deletion (746 to 750 residues) mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of wild type EGFR (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of EGFR T790M/L858R mutant (unknown origin) by high-throughput biochemical screeningMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.100nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of recombinant human N-terminal GST-tagged EGFR catalytic domain (669 to 1210 residues) expressed in baculovirus expression system by mass...More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of wild-type EGFR (unknown origin) by high-throughput biochemical screeningMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.107nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.150nMAssay Description:Binding affinity to recombinant Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substra...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.150nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.157nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.188nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Inhibition of EGFR deletion (746 to 750 residues) mutant (unknown origin)More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.289nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.300nMAssay Description:Inhibition of EGFR T790M/del746 to 750 mutant (unknown origin) by high-throughput biochemical screeningMore data for this Ligand-Target Pair
Affinity DataKi: 0.300nMAssay Description:Inhibition of EGFR L858R mutant (unknown origin)More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.320nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.323nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.346nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.350nMAssay Description:Inhibition of human EGFR T790M/del (746 to 750 residues) mutant assessed as apparent inhibition constant using Fl-EEPLYWSFPAKKK-CONH2 peptide as subs...More data for this Ligand-Target Pair
Ligand Info
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.355nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.380nMAssay Description:Competitive inhibition of Aurora B ATP binding siteMore data for this Ligand-Target Pair
Affinity DataKi: 0.380nMAssay Description:Competitive inhibition of human Aurora B ATP binding site by rapid dilution methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.400nMAssay Description:Inhibition of EGFR deletion (746 to 750 residues) mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.420nMAssay Description:Binding affinity to recombinant Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substra...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.440nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.5nMAssay Description:Inhibition of EGFR L858R mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: <0.5nMAssay Description:Inhibition of EGFR deletion (746 to 750 residues) mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: <0.5nMAssay Description:Inhibition of EGFR L858R mutant (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.510nMAssay Description:Binding affinity to recombinant Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substra...More data for this Ligand-Target Pair
Affinity DataKi: 0.510nMAssay Description:Binding affinity to recombinant Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substra...More data for this Ligand-Target Pair
Affinity DataKi: >0.530nMAssay Description:Inhibition of human wild type EGFR using Fl-EEPLYWSFPAKKK-CONH2 as substrate preincubated for 30 mins followed by addition of substrate measured afte...More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.560nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: <0.600nMAssay Description:Inhibition of human EGFR T790M/L858R double mutant assessed as apparent inhibition constant using Fl-EEPLYWSFPAKKK-CONH2 as peptide substrate preincu...More data for this Ligand-Target Pair
Ligand Info
Affinity DataKi: <0.600nMAssay Description:Inhibition of human EGFR T790M/L858R double mutant assessed as apparent inhibition constant using Fl-EEPLYWSFPAKKK-CONH2 as peptide substrate preincu...More data for this Ligand-Target Pair
Ligand Info
Affinity DataKi: 0.600nMAssay Description:Inhibition of Aurora AMore data for this Ligand-Target Pair
Affinity DataKi: 0.600nMAssay Description:Inhibition of wild type EGFR (unknown origin)More data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.681nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
TargetPhosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform(Homo sapiens (Human))
Genentech
Curated by ChEMBL
Genentech
Curated by ChEMBL
Affinity DataKi: 0.692nMAssay Description:Inhibition of PI3Kalpha (unknown origin) using PIP2:3PS as substrate in presence of ATP measured after 120 mins by ADP-Glo assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.780nMAssay Description:Binding affinity to recombinant Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substra...More data for this Ligand-Target Pair



 3D Structure (crystal)
3D Structure (crystal)