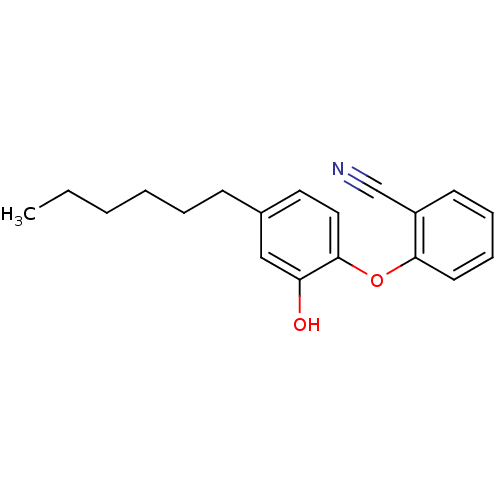
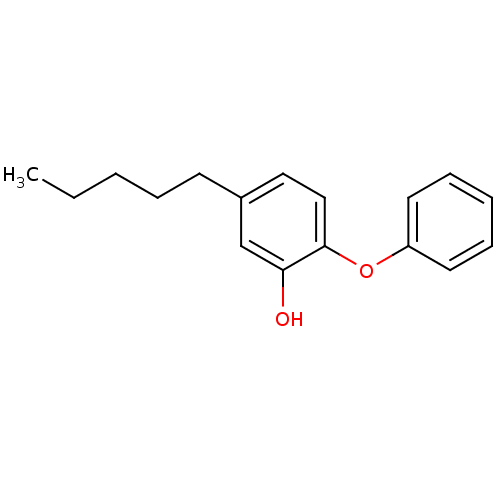
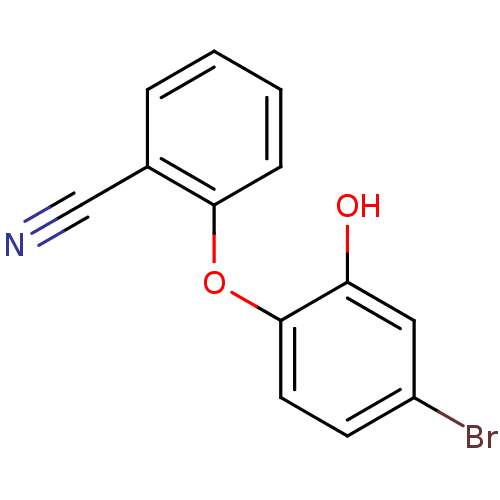
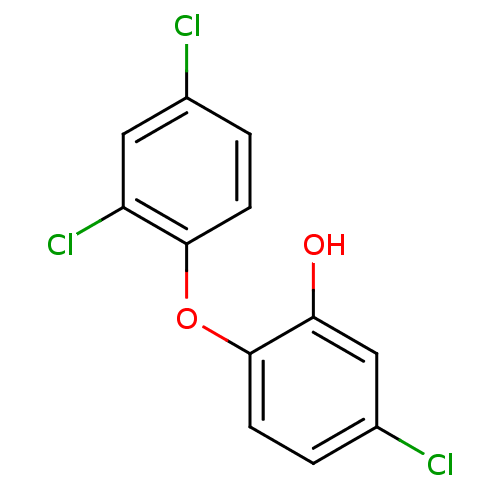
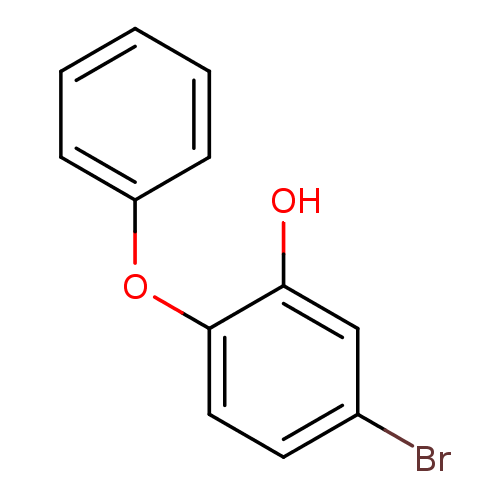
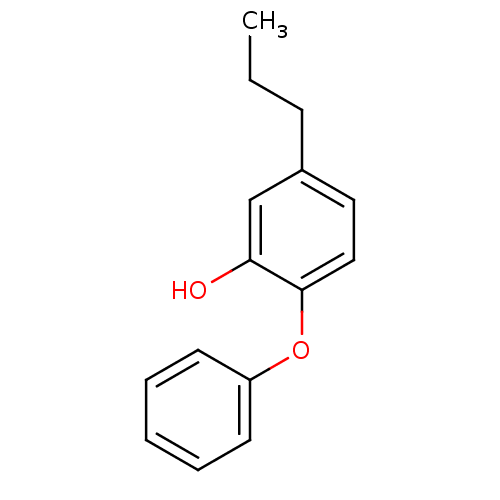
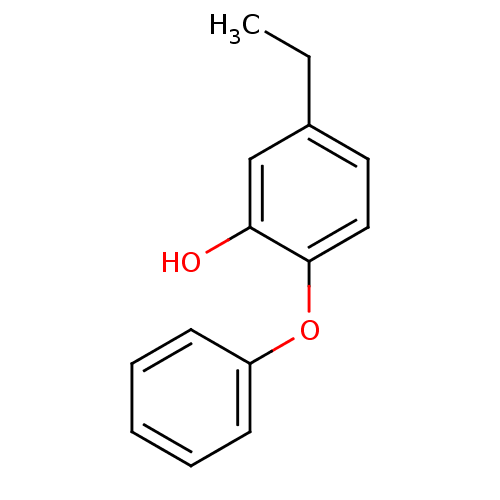
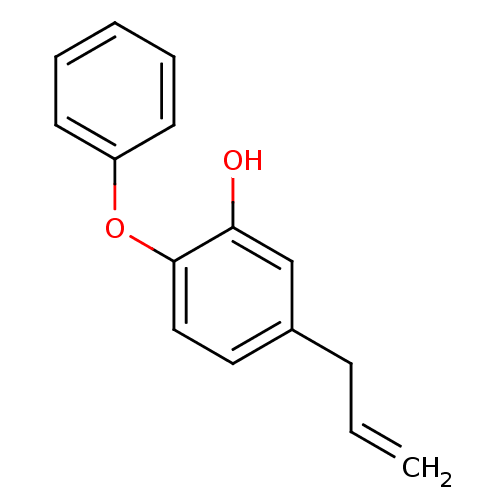
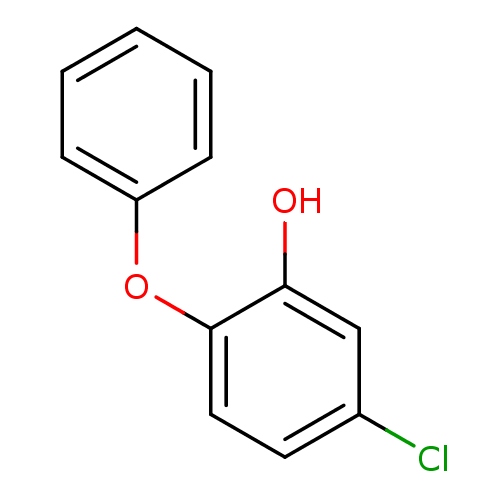
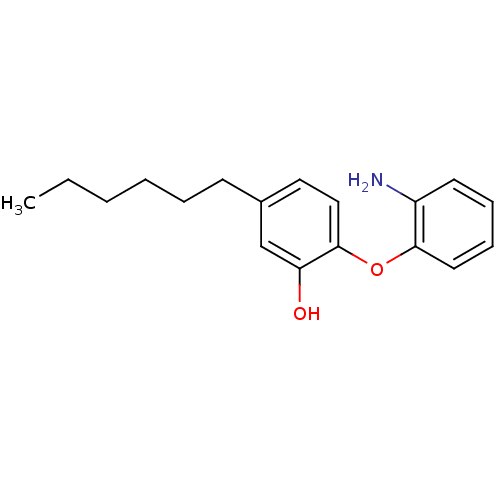
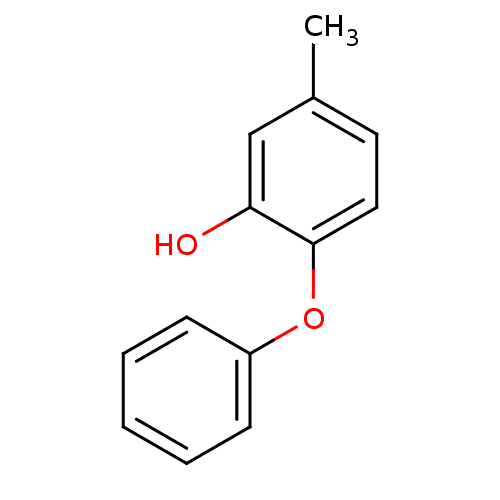
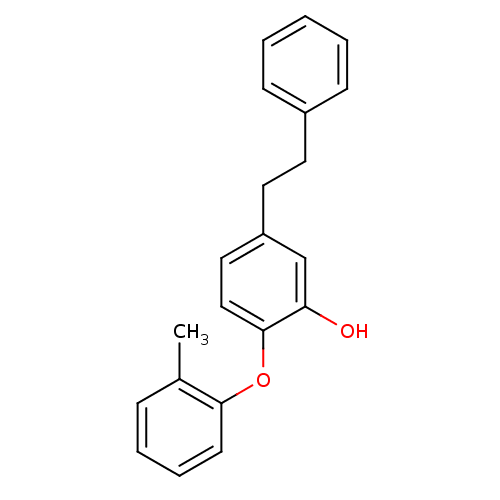
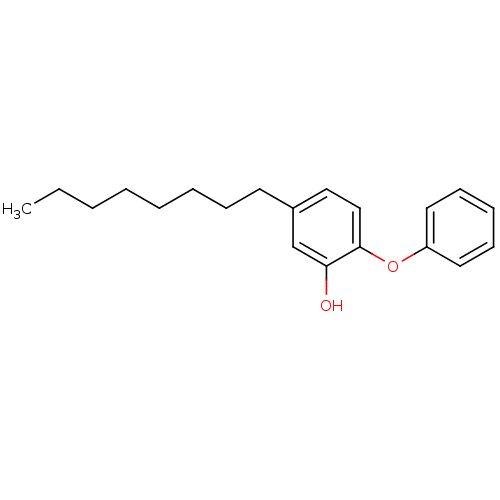
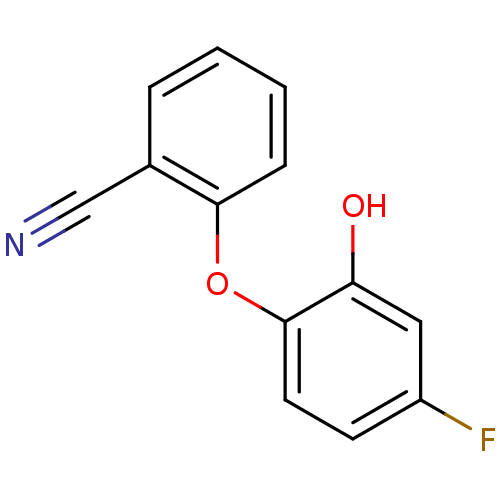
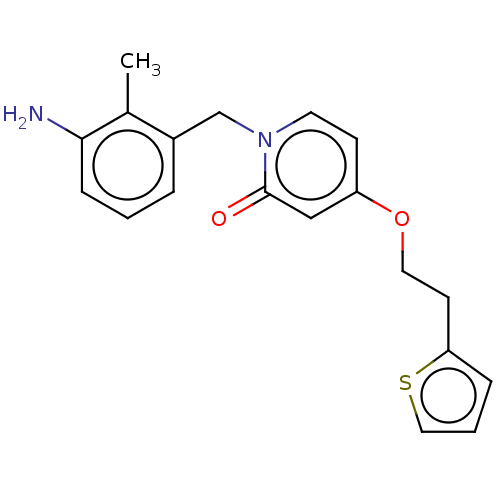
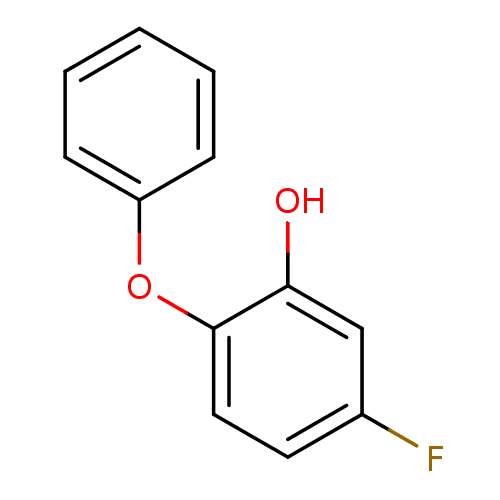
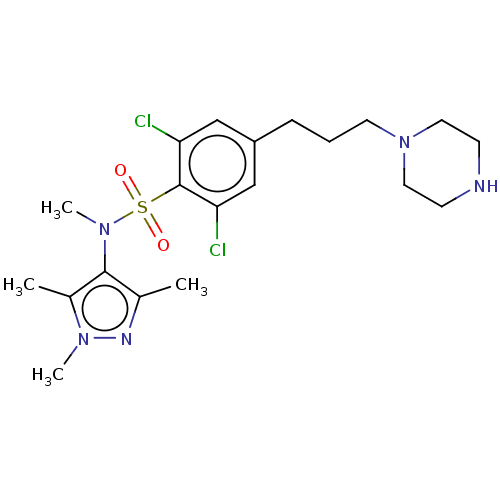
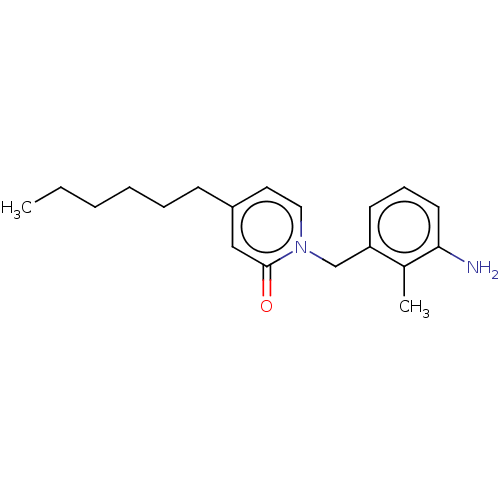
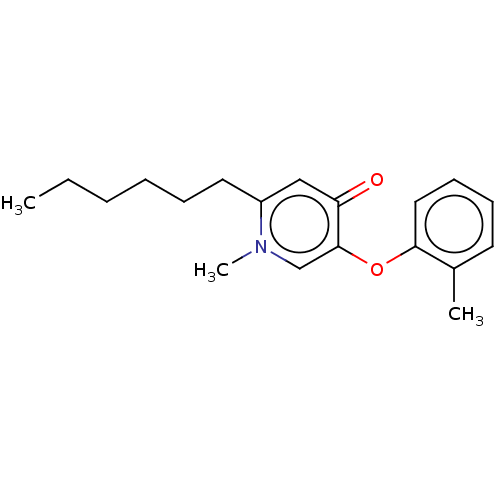
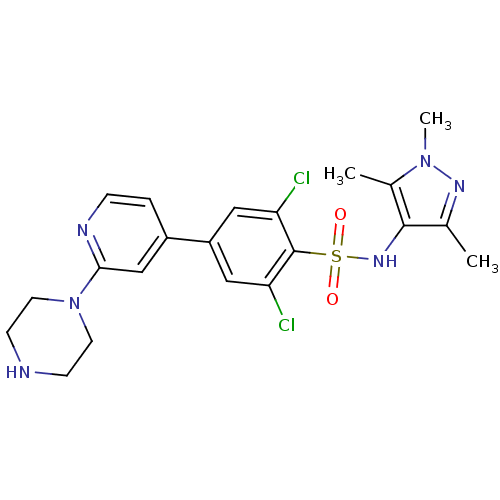
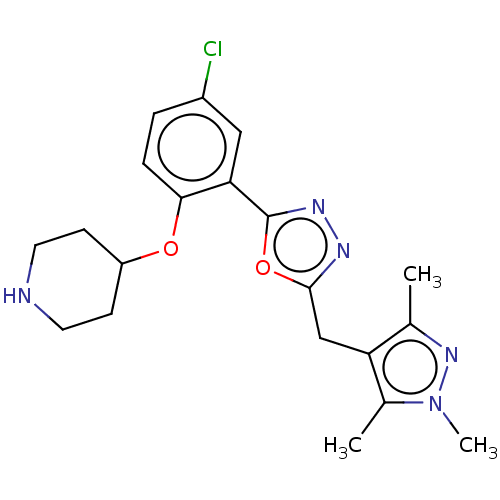
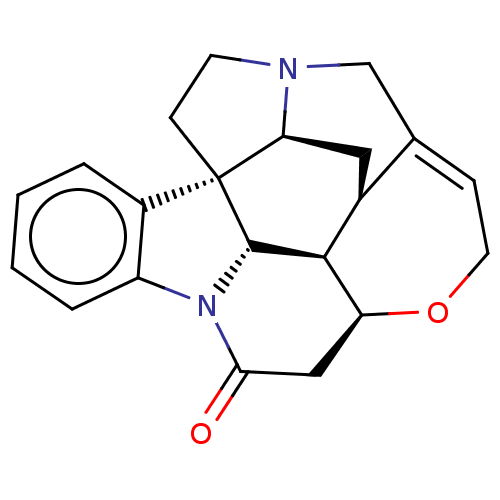
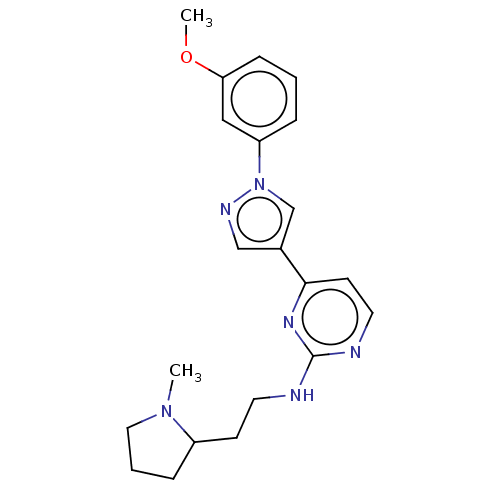
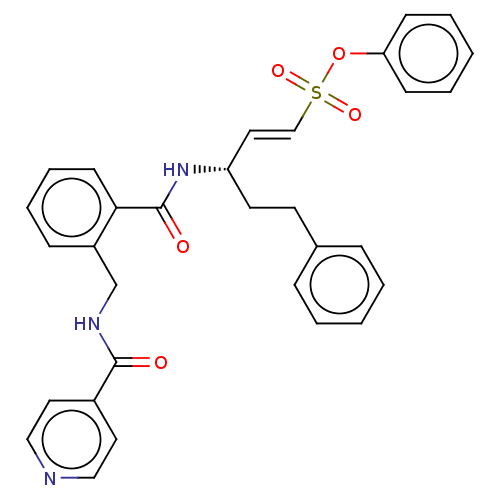
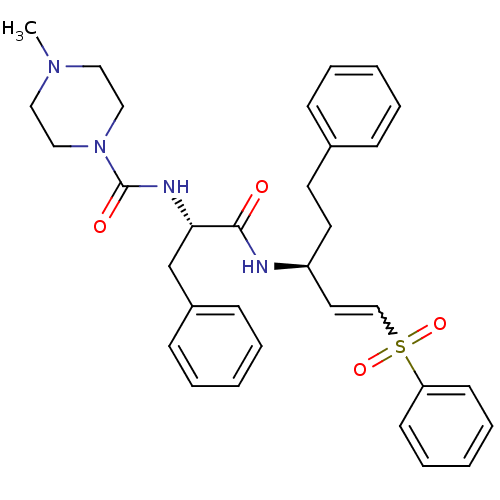
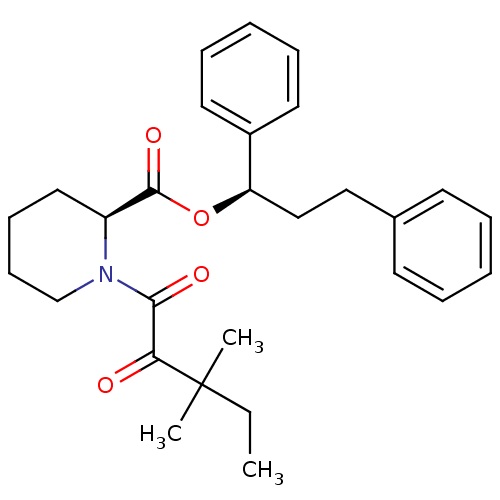
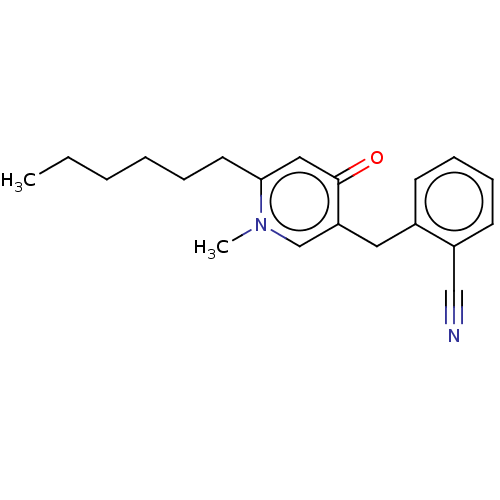
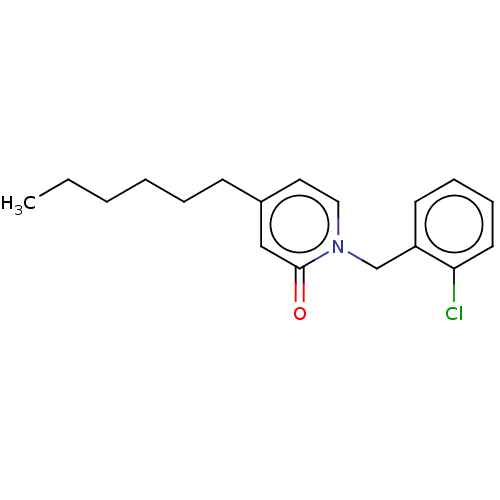
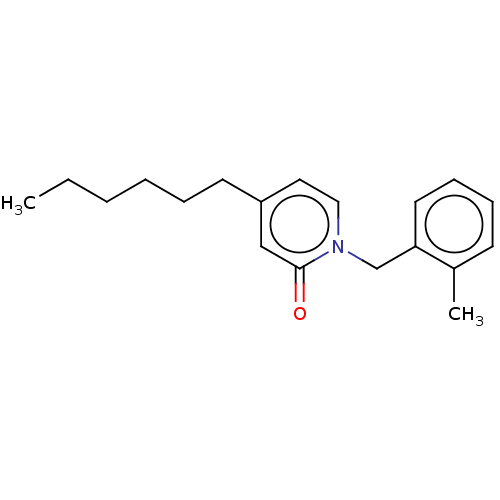
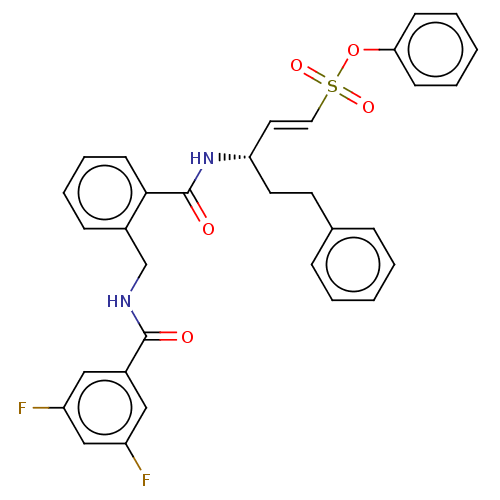
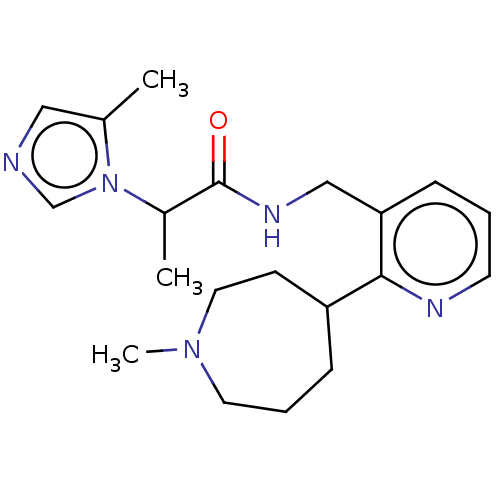
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0400nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0500nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0500nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0600nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0600nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0700nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.0900nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.100nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.110nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.120nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.120nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.380nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.5nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 0.820nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 0.900nMAssay Description:Inhibition of human NMT1 L495M mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after 30 mins b...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.01nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.30nMAssay Description:Inhibition of Leishmania major NMT H398N mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 1.30nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.40nMAssay Description:Inhibition of N-terminal TEV cleavage site-fused His6 tagged Leishmania major NMT (11 to 421 residues) expressed in Escherichia coli Rosetta-2 cells ...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.42nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
Stony Brook University
Stony Brook University
Affinity DataKi: 1.49nMpH: 7.5Assay Description:ThermoFluor experiments were carried out in 96-well plates (Concord) using the CFX96 RealTime PCR Detection system and C1000 Thermal Cycler (BioRad).More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 1.60nMAssay Description:Inhibition of Leishmania major NMT H398N mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 2nMAssay Description:Inhibition of N-terminal TEV cleavage site-fused His6 tagged Leishmania major NMT (11 to 421 residues) expressed in Escherichia coli Rosetta-2 cells ...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 2nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 2.30nMAssay Description:Inhibition of Leishmania major NMT L421Q mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 2.70nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 4nMAssay Description:Inhibition of human NMT1 L495M mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after 30 mins b...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 4.80nMAssay Description:Inhibition of human NMT1 R295Q/W297F/A452M/L453V/L462V/N473H/L495M/Q496L mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as subst...More data for this Ligand-Target Pair
TargetGlycine receptor subunit alpha-1(Homo sapiens (Human))
The German University In Cairo
Curated by ChEMBL
The German University In Cairo
Curated by ChEMBL
Affinity DataKi: 5nMAssay Description:Displacement of [3H]strychnine from human glycine receptor subunit alpha-1 expressed in HEK293 cell membranes preincubated for 30 mins followed by [3...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 6nMAssay Description:Inhibition of Leishmania major NMT L421Q mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 6.20nMAssay Description:Inhibition of human NMT1 N473H/L495M/Q496L mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA aft...More data for this Ligand-Target Pair
Affinity DataKi: 7nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 7.30nMAssay Description:Inhibition of N-terminal TEV cleavage site-fused His6 tagged Leishmania major NMT (11 to 421 residues) expressed in Escherichia coli Rosetta-2 cells ...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 8.40nMAssay Description:Inhibition of N-terminal TEV cleavage site-fused His6 tagged Leishmania major NMT (11 to 421 residues) expressed in Escherichia coli Rosetta-2 cells ...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 8.5nMAssay Description:Inhibition of Leishmania major NMT H398N mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg University
Curated by ChEMBL
Johannes Gutenberg University
Curated by ChEMBL
Affinity DataKi: 9nMAssay Description:Irreversible inhibition of Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substrate by...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:Irreversible inhibition of human Cathepsin L assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substrate by fluorometric assayMore data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg University
Curated by ChEMBL
Johannes Gutenberg University
Curated by ChEMBL
Affinity DataKi: 10nMAssay Description:Irreversible inhibition of Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substrate by...More data for this Ligand-Target Pair
TargetPeptidyl-prolyl cis-trans isomerase FKBP1A(Homo sapiens (Human))
University Of Wu£Rzburg
Curated by ChEMBL
University Of Wu£Rzburg
Curated by ChEMBL
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 11nMAssay Description:Inhibition of human NMT1 W297F/A452M/L453V/L462V/L495M/Q496L mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in pres...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 11nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 11nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
TargetEnoyl-[acyl-carrier-protein] reductase [NADPH] FabI(Staphylococcus aureus)
University of Wuerzburg
University of Wuerzburg
Affinity DataKi: 11.9nMpH: 7.5Assay Description:Kinetics were performed on a Cary 100 spectrophotometer (Varian) at 20 °C. Reaction velocities were measured by monitoring the oxidation of NAD(P)H t...More data for this Ligand-Target Pair
Affinity DataKi: 12nMAssay Description:Displacement of [3H]strychnine from glycine receptor alpha 1 in rat brain synaptic membranesMore data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 13nMAssay Description:Inhibition of N-terminal TEV cleavage site-fused His6 tagged human NMT1 (115 to 496 residues) expressed in Escherichia coli Rosetta-2 cells using GSN...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase(Leishmania major)
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 14nMAssay Description:Inhibition of Leishmania major NMT H398N mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as substrate in presence of MyrCoA after...More data for this Ligand-Target Pair
TargetCysteine protease(Trypanosoma brucei rhodesiense)
Johannes Gutenberg University
Curated by ChEMBL
Johannes Gutenberg University
Curated by ChEMBL
Affinity DataKi: 15nMAssay Description:Irreversible inhibition of Trypanosoma brucei rhodesiense rhodesain assessed as inhibition constant using Cbz-Phe-Arg-AMC as fluorogenic substrate by...More data for this Ligand-Target Pair
TargetGlycylpeptide N-tetradecanoyltransferase 1(Homo sapiens (Human))
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Johannes Gutenberg-Universit£T Mainz
Curated by ChEMBL
Affinity DataKi: 15nMAssay Description:Inhibition of human NMT1 R295Q/W297F/A452M/L453V/L462V/N473H/L495M/Q496L mutant expressed in Escherichia coli Rosetta-2 cells using GSNKSKPK as subst...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)