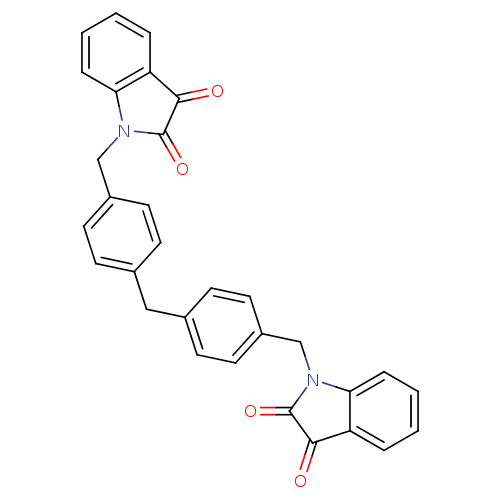
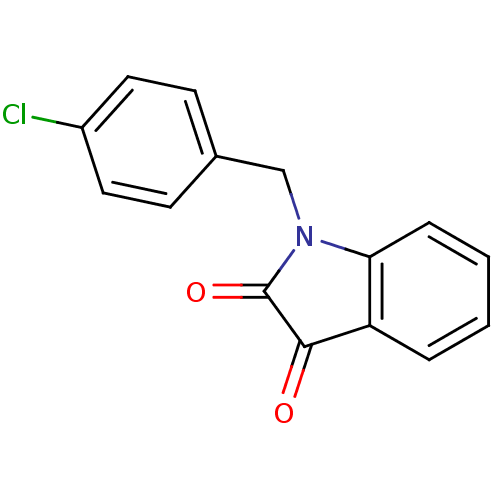
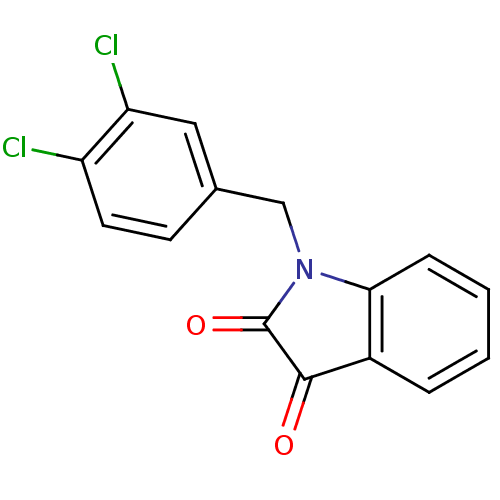
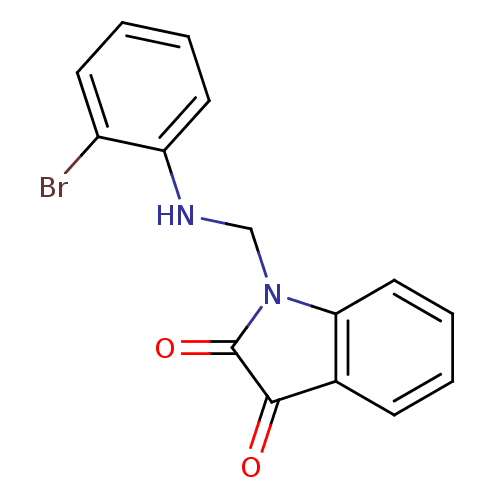
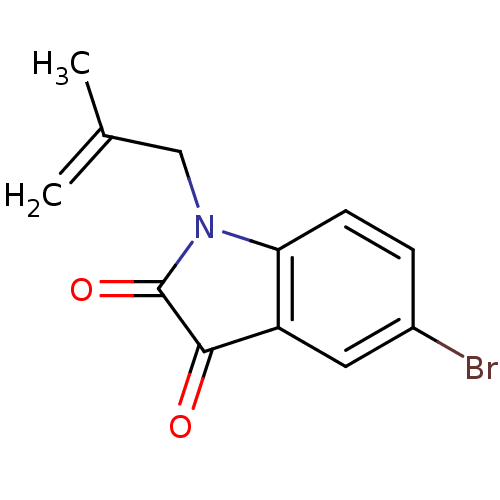
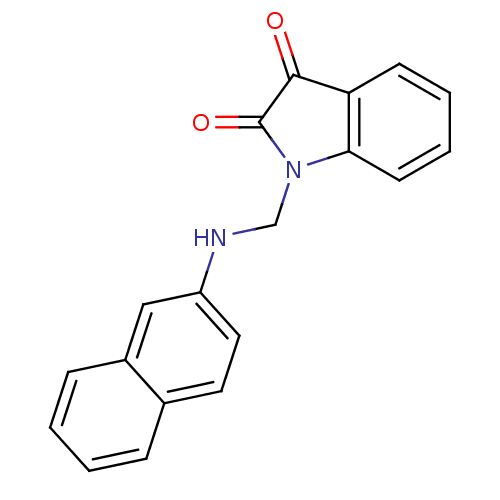
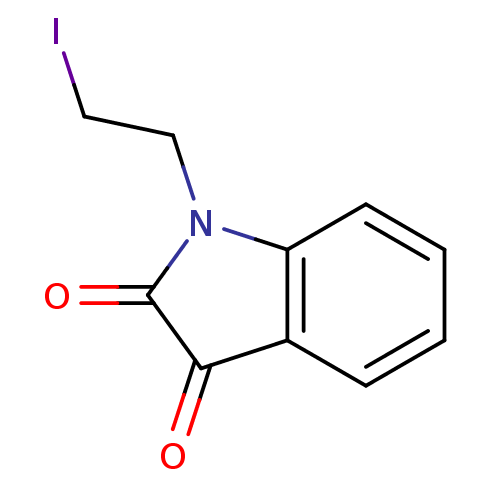
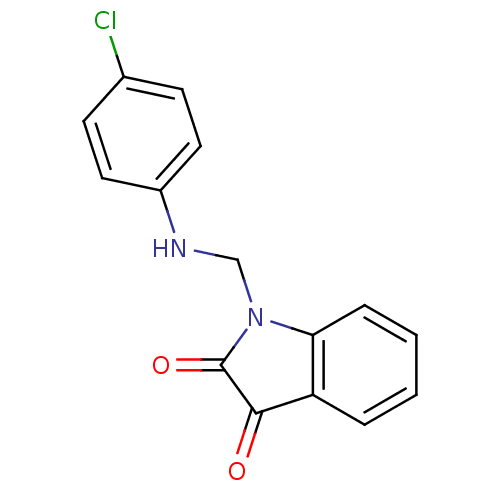
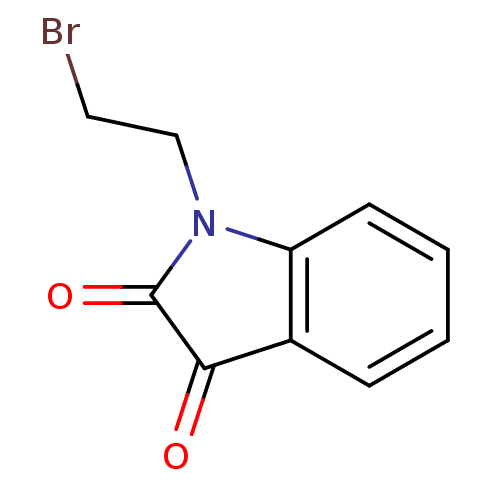
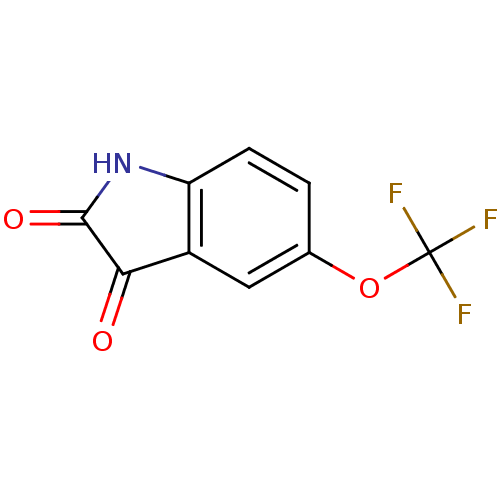
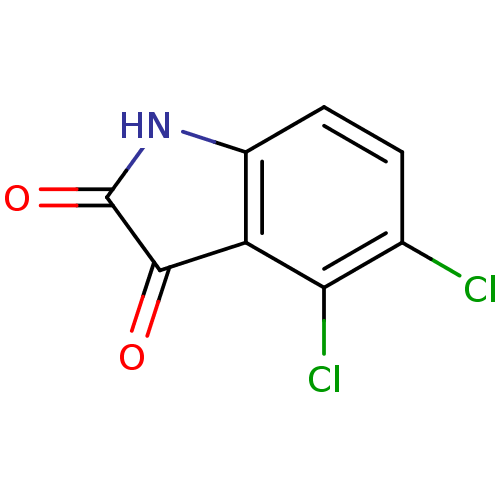
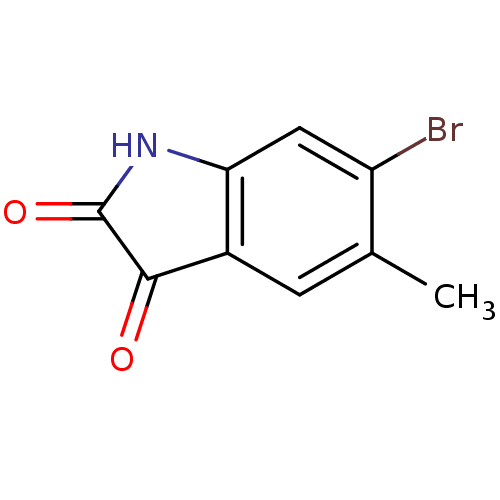
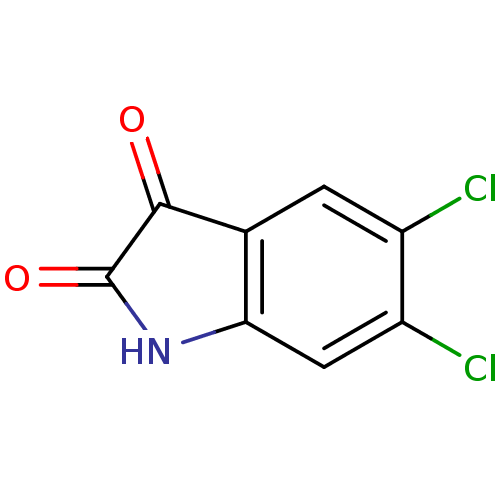
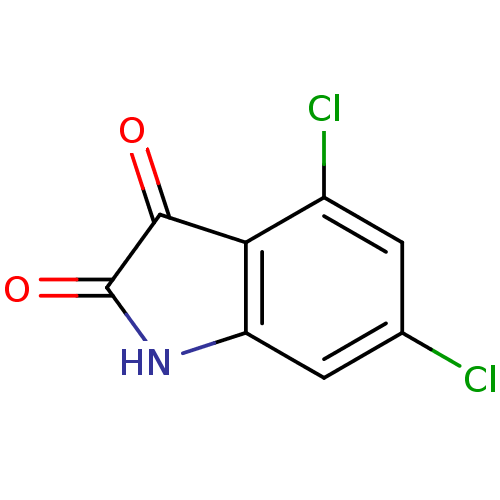
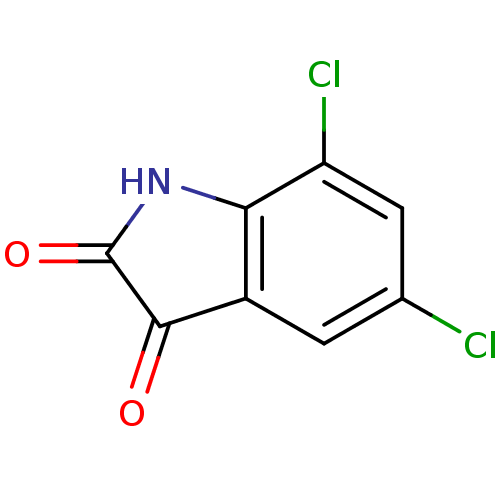
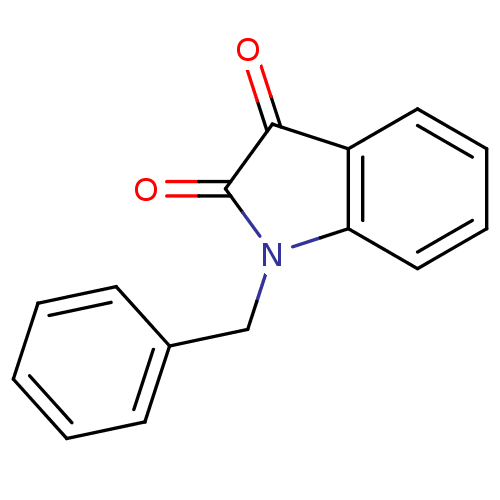
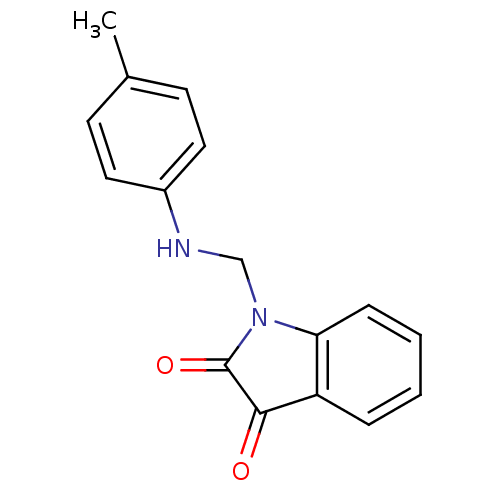
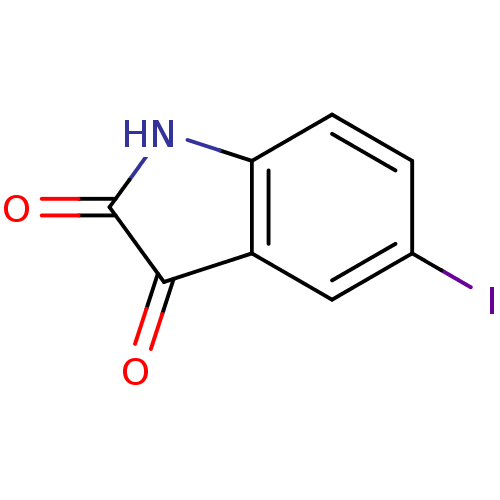
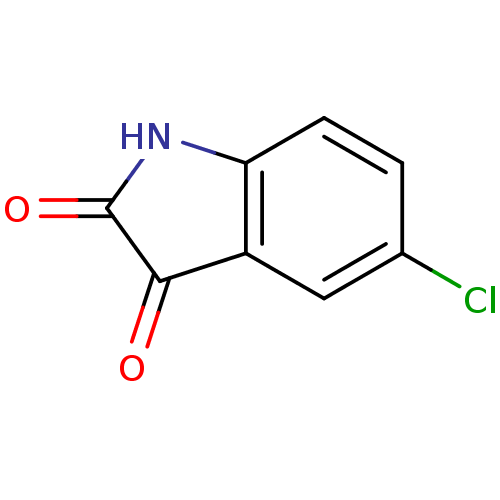
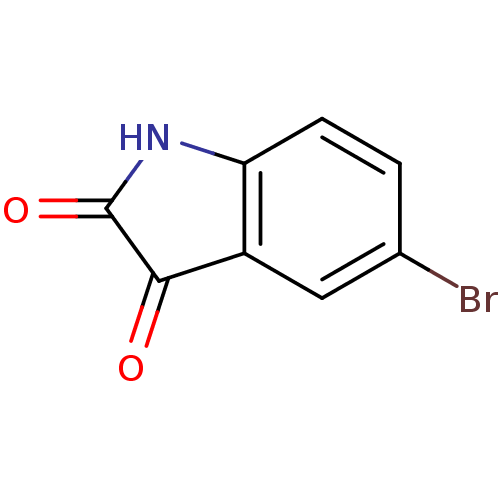
Affinity DataKi: 6nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 8nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 8nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 10nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 16nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 23nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 25nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 31nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 32nM ΔG°: -42.8kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 47nM ΔG°: -41.8kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 65nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 66nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 67nM ΔG°: -40.9kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 88nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 110nM ΔG°: -39.7kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 170nM ΔG°: -38.6kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 200nM ΔG°: -38.2kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 280nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 290nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 360nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 410nM ΔG°: -36.5kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 420nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 490nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 530nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 580nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 590nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 610nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 610nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 610nM ΔG°: -35.5kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 620nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 650nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 740nM ΔG°: -35.0kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 750nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 760nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 870nM ΔG°: -34.6kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 890nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 950nM ΔG°: -34.4kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.08E+3nM ΔG°: -34.1kJ/molepH: 7.4 T: 2°CAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.12E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.15E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.16E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.22E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.25E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair
Affinity DataKi: 1.32E+3nMAssay Description:CE inhibition was determined using a spectrophotometric multiwell plate assay with o-NPA as a substrate. The rate of change in absorbance at 420 nm w...More data for this Ligand-Target Pair