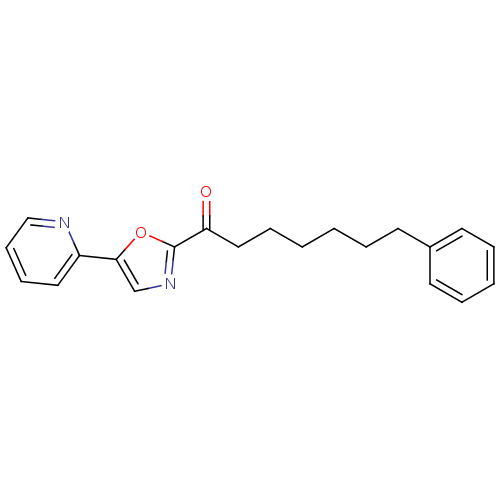
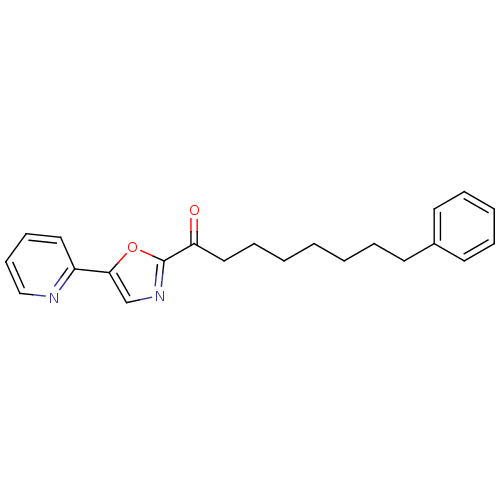
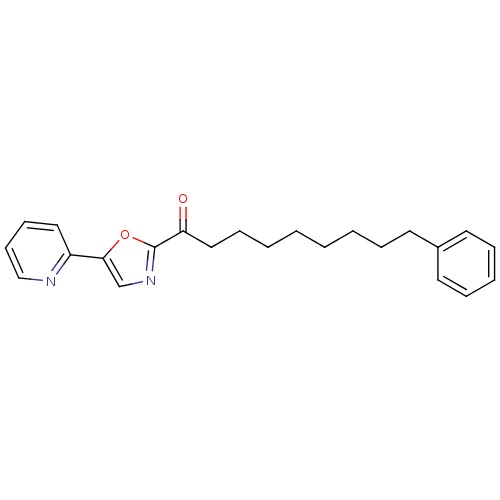
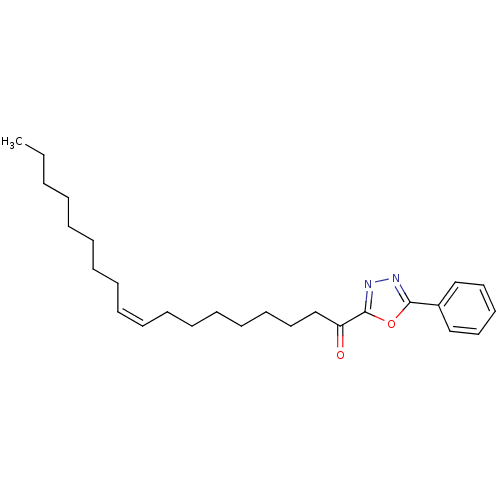
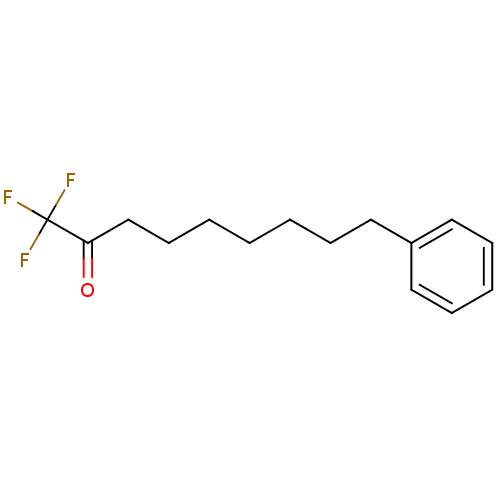
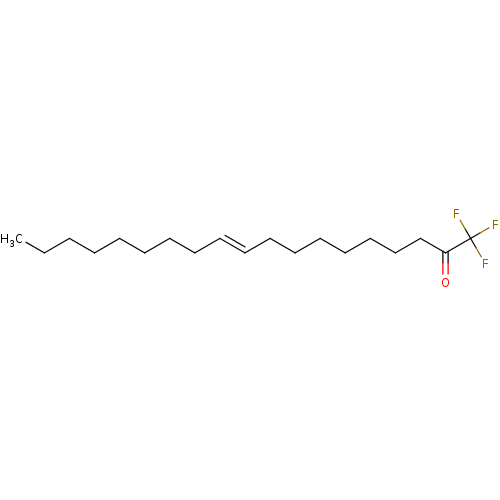
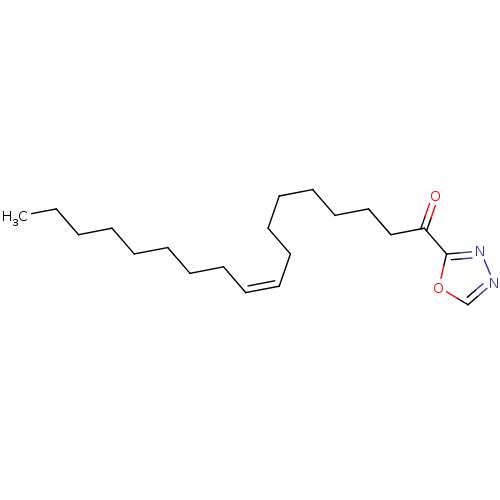
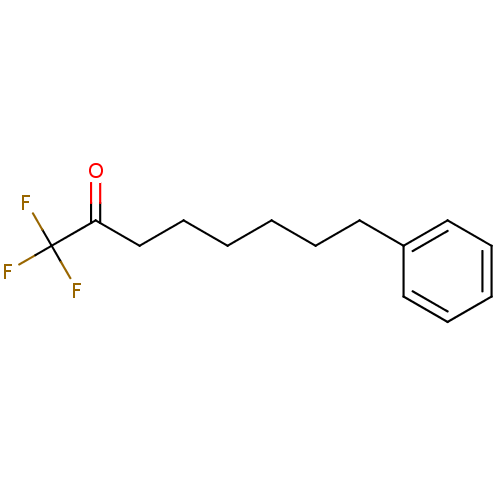
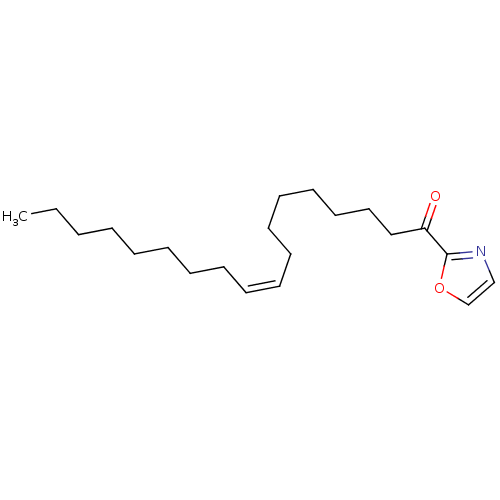
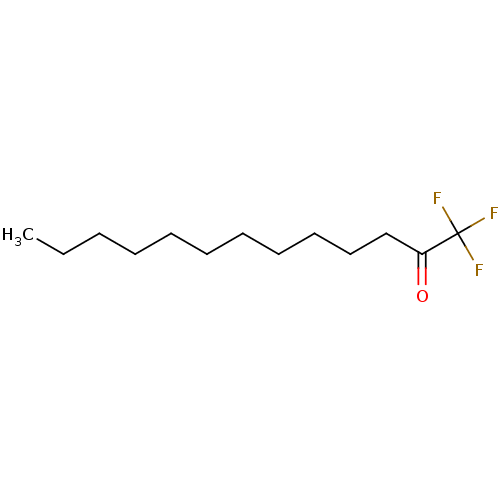
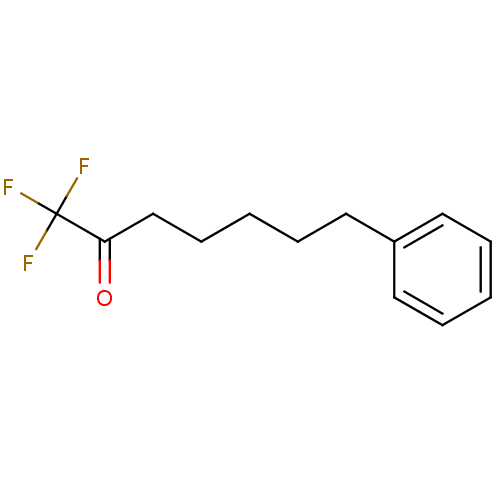
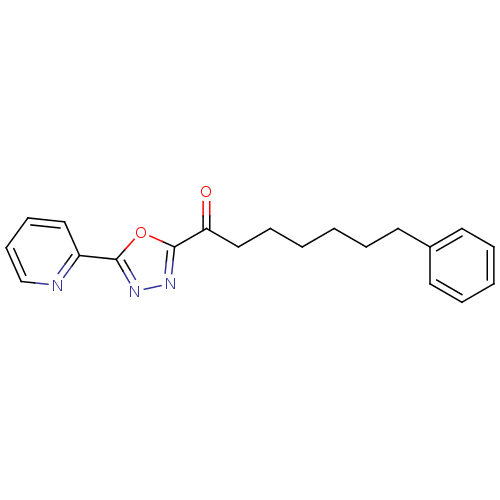
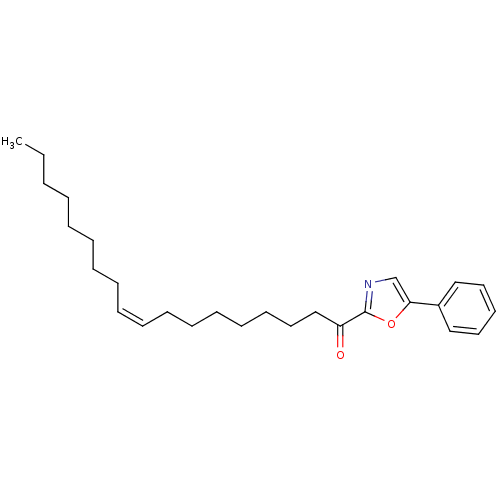
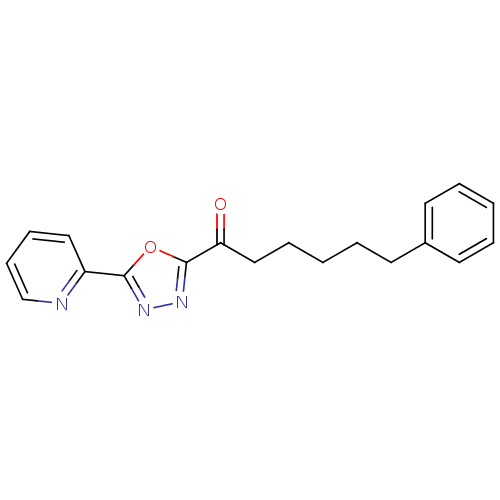
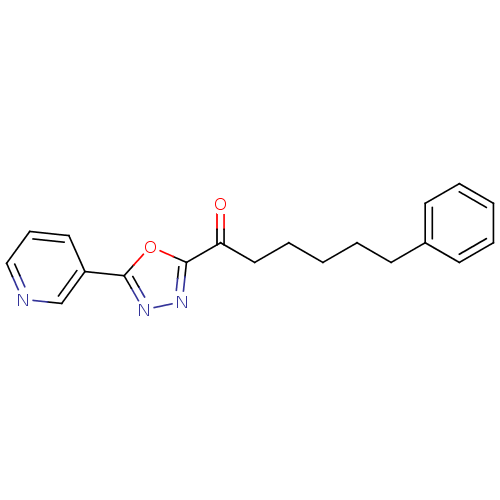
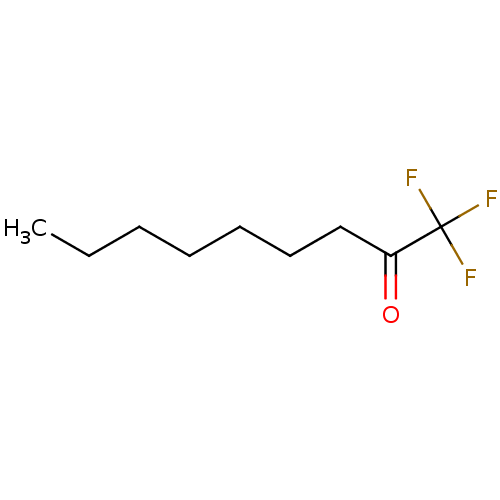
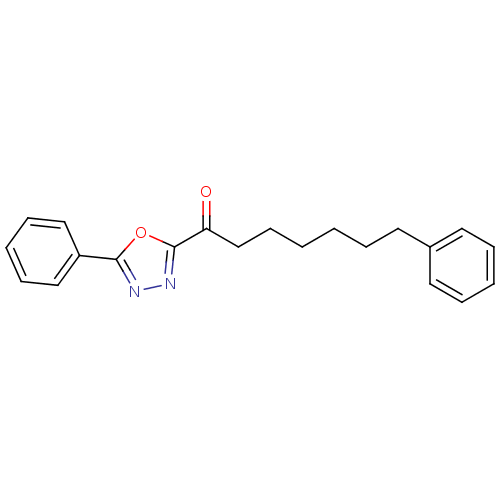
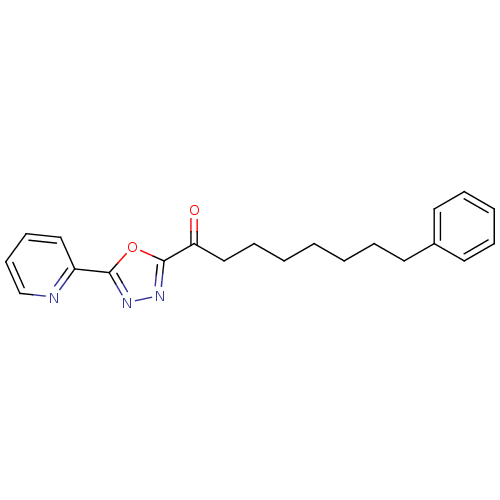
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.200nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.280nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.290nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.390nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.520nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.560nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.830nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 0.850nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 1nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 1nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 2.30nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 3nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 4.70nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 7.5nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 7.80nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 11nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 16nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 18nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 25nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 80nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 90nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 100nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 100nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 130nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 140nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 170nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 220nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 240nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 290nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 290nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 320nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 370nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 560nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 560nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 850nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 1.20E+3nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 2.00E+3nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 3.20E+3nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Mus musculus (mouse))
The Scripps Research Institute
Curated by ChEMBL
The Scripps Research Institute
Curated by ChEMBL
Affinity DataKi: 3.20E+3nMAssay Description:Inhibitor affinity of the compound towards enzymes of class serine hydrolase was determined using biotin or fluorescent as radioligand (FP-biotin or ...More data for this Ligand-Target Pair

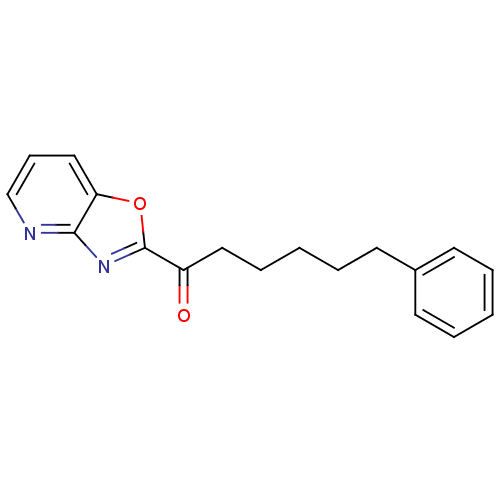

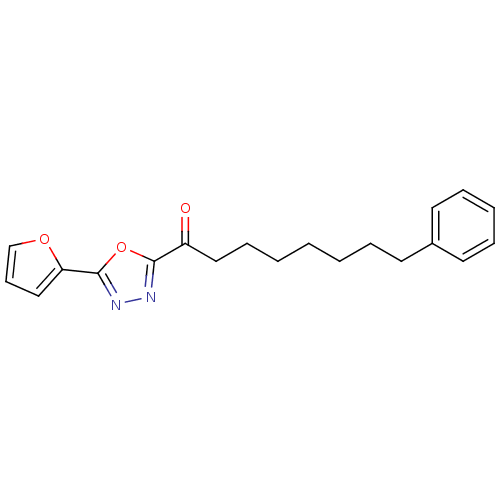
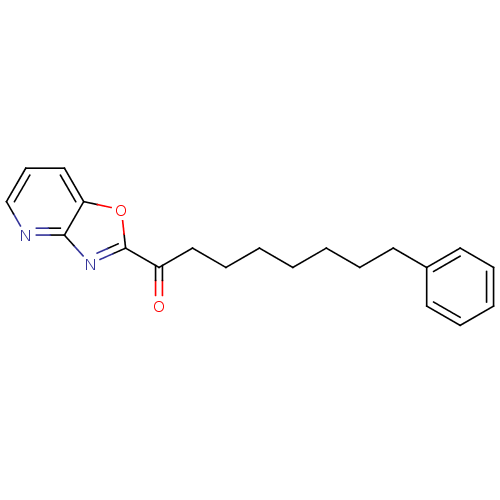
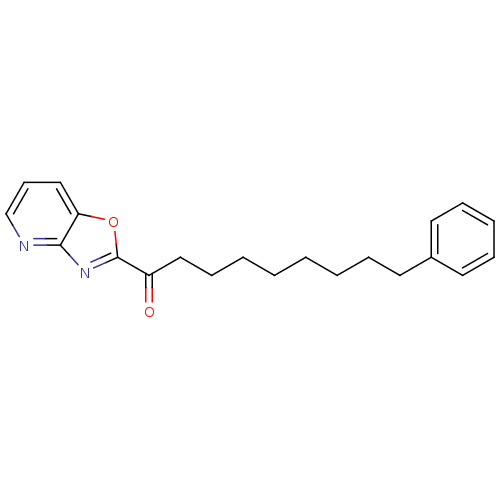

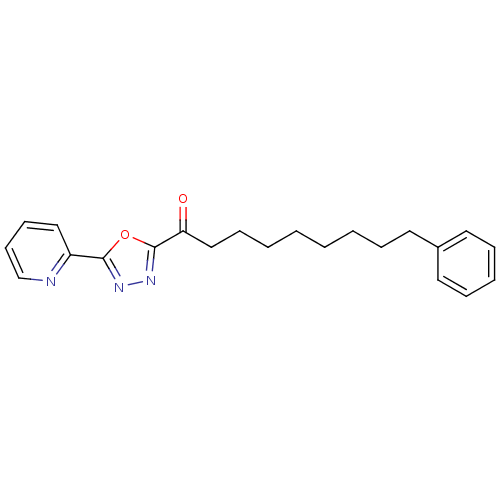
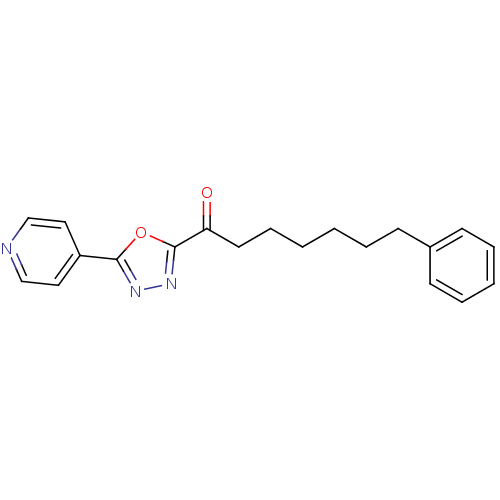
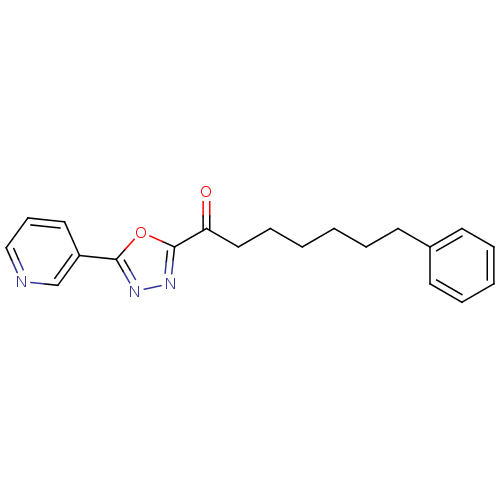
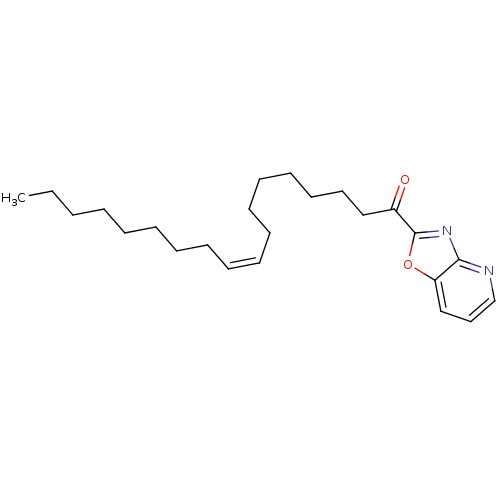

 3D Structure (crystal)
3D Structure (crystal)