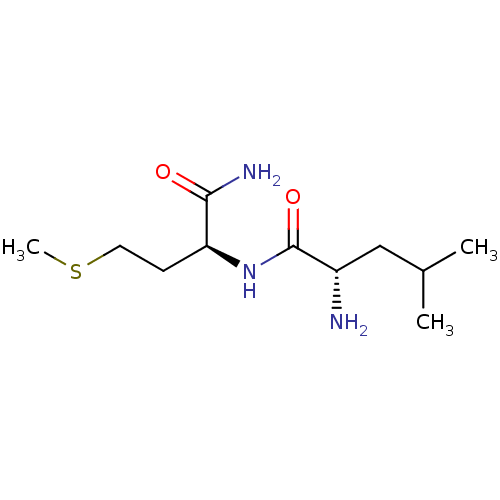
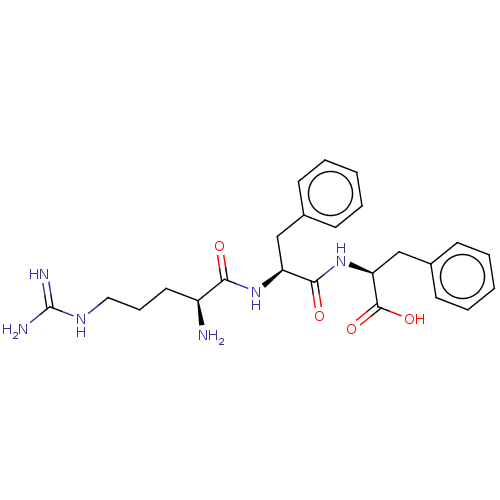
TargetGeneral amino-acid permease GAP1(Saccharomyces cerevisiae)
Katholieke Universiteit Leuven
Curated by ChEMBL
Katholieke Universiteit Leuven
Curated by ChEMBL
Affinity DataKi: 1.05E+5nMAssay Description:Competitive inhibition of nitrogen-starved wild type sigma1278b yeast Gap1 by Lineweaver-Burke plot analysisMore data for this Ligand-Target Pair
TargetGeneral amino-acid permease GAP1(Saccharomyces cerevisiae)
Katholieke Universiteit Leuven
Curated by ChEMBL
Katholieke Universiteit Leuven
Curated by ChEMBL
Affinity DataKi: 1.90E+5nMAssay Description:Competitive inhibition of nitrogen-starved wild type sigma1278b yeast Gap1 by Lineweaver-Burke plot analysisMore data for this Ligand-Target Pair
TargetGeneral amino-acid permease GAP1(Saccharomyces cerevisiae)
Katholieke Universiteit Leuven
Curated by ChEMBL
Katholieke Universiteit Leuven
Curated by ChEMBL
Affinity DataKi: 5.00E+6nMAssay Description:Noncompetitive inhibition of nitrogen-starved wild type sigma1278b Gap1-mediated amino acid transport by Lineweaver-Burke plot analysisMore data for this Ligand-Target Pair
TargetGeneral amino-acid permease GAP1(Saccharomyces cerevisiae)
Katholieke Universiteit Leuven
Curated by ChEMBL
Katholieke Universiteit Leuven
Curated by ChEMBL
Affinity DataKi: 1.20E+7nMAssay Description:Noncompetitive inhibition of nitrogen-starved wild type sigma1278b Gap1-mediated amino acid transport by Lineweaver-Burke plot analysisMore data for this Ligand-Target Pair