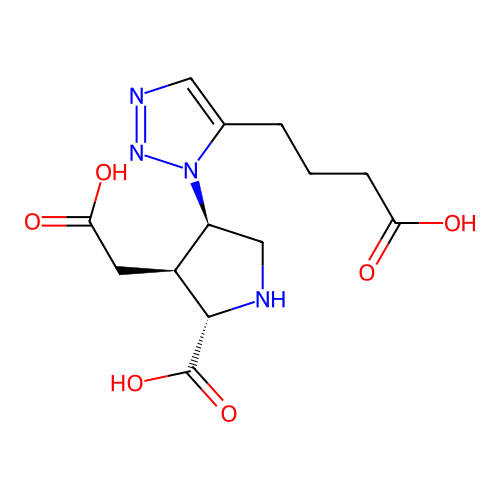
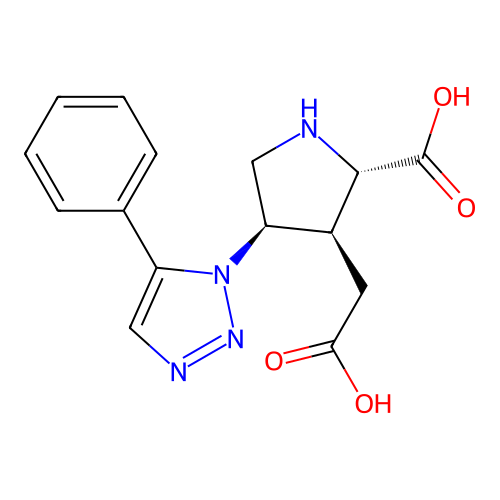
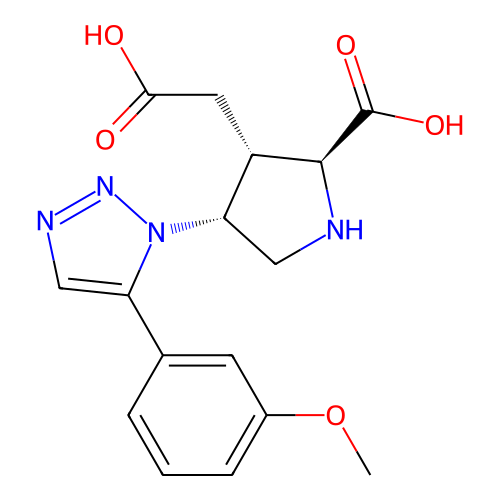
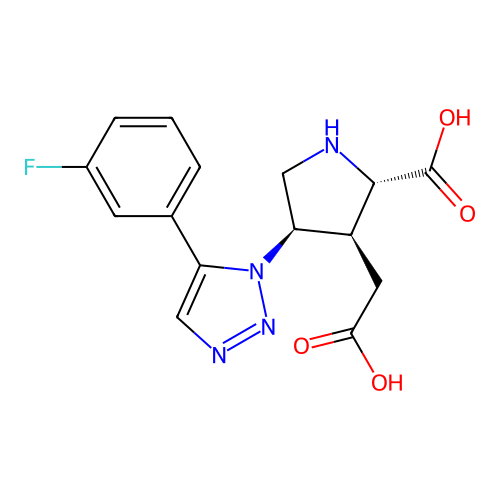
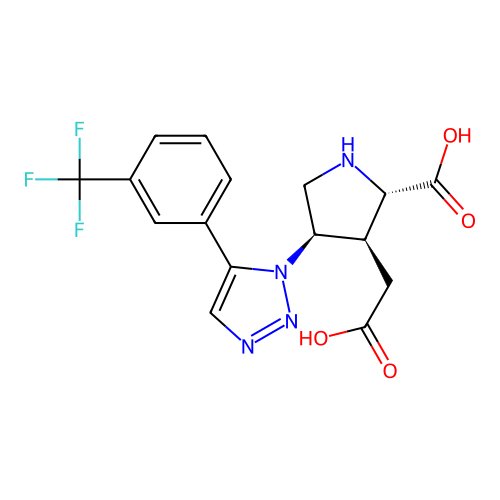
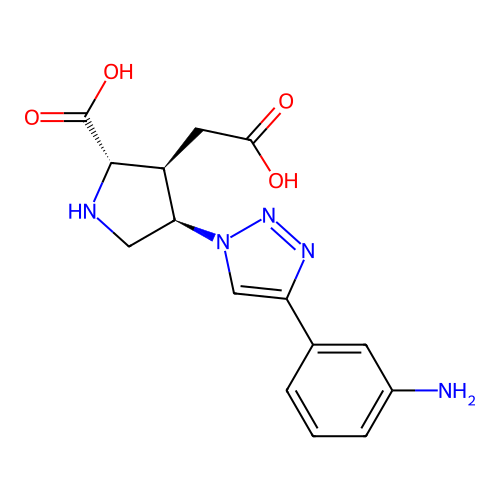
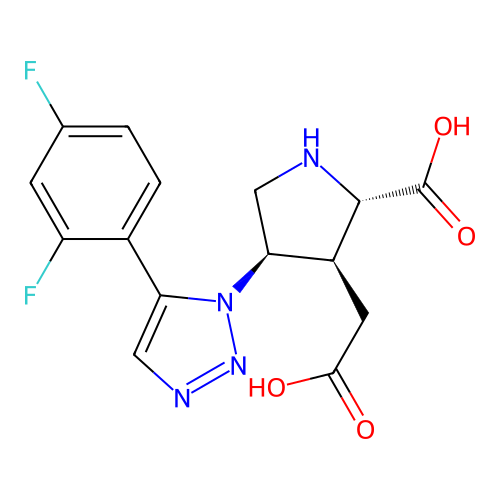
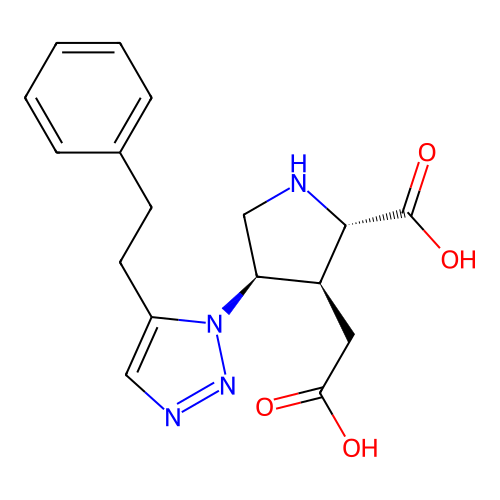
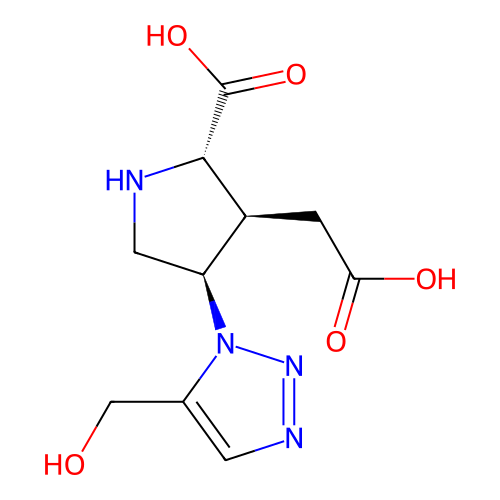
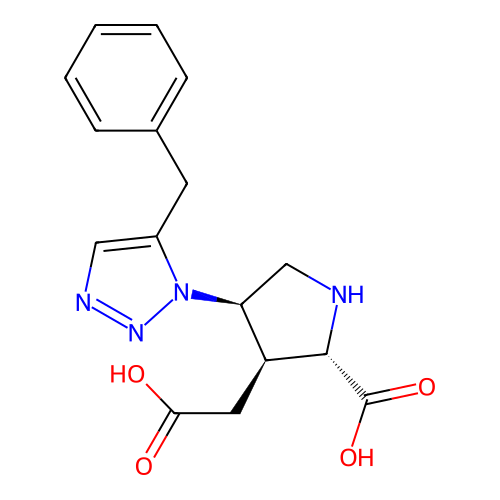
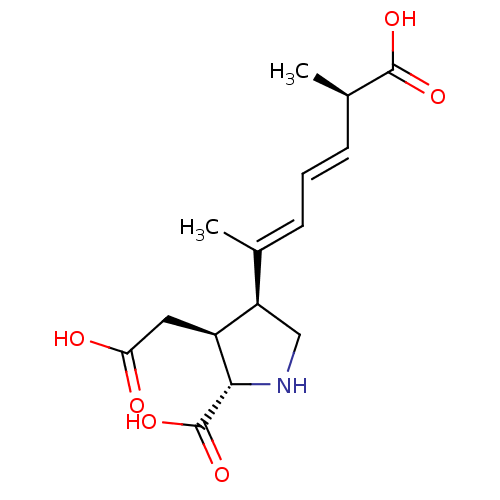
Report error Found 49 Enz. Inhib. hit(s) with all data for entry = 50021916
Affinity DataKi: 1.10nMAssay Description:Displacement of [3H](2S, 4R)-4-MeGlu (SYM-2081) from GluK1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 4.80nMAssay Description:Displacement of [3H](2S, 4R)-4-MeGlu (SYM-2081) from GluK3 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 6nMAssay Description:Displacement of [3H](2S, 4R)-4-MeGlu (SYM-2081) from GluK2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat synaptic membraneMore data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:Displacement of [3H]-kainic acid from GluR6 (unknown origin) expressed in Sf9 cells membraneMore data for this Ligand-Target Pair
Affinity DataIC50: 130nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat synaptic membraneMore data for this Ligand-Target Pair
Affinity DataKi: 258nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 280nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataKi: 420nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 432nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataIC50: 470nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat synaptic membraneMore data for this Ligand-Target Pair
Affinity DataKi: 483nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 499nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.02E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.76E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+3nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat synaptic membraneMore data for this Ligand-Target Pair
Affinity DataKi: 2.15E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 2.60E+3nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataKi: 3.25E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 3.30E+3nMAssay Description:Displacement of [3H]-NF608 from rat GluK1More data for this Ligand-Target Pair
Affinity DataIC50: 3.60E+3nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataKi: 3.75E+3nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataKi: 5.35E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 5.57E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+3nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataKi: 5.90E+3nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataKi: 6.10E+3nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataKi: 7.76E+3nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataKi: 7.79E+3nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataKi: 7.97E+3nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 8.66E+3nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataIC50: 9.00E+3nMAssay Description:Binding affinity to AMPA receptor in rat synaptosomesMore data for this Ligand-Target Pair
Affinity DataIC50: 9.50E+3nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat cortical synaptosomes incubated for 30 mins by TopCount microplate scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.08E+4nMAssay Description:Displacement of [3H]-NF608 from rat GluK1More data for this Ligand-Target Pair
Affinity DataKi: 1.11E+4nMAssay Description:Displacement of [3H]-kainate from mouse GluK5 expressed in Sf9 cell membrane incubated for 2 hrs by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 1.15E+4nMAssay Description:Displacement of [3H]-NF608 from rat GluK1More data for this Ligand-Target Pair
Affinity DataKi: 1.17E+4nMAssay Description:Displacement of [3H]-NF608 from rat GluK1More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+4nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataIC50: 1.70E+4nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataKi: 1.79E+4nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataKi: 2.11E+4nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataIC50: 3.20E+4nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataKi: 3.60E+4nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataKi: 7.00E+4nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]-KA from rat GluK3More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat cortical synaptosomes incubated for 30 mins by TopCount microplate scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]-NF608 from rat GluK1More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Displacement of [3H]-AMPA from AMPA in rat brain membrane preincubated for 15 mins followed by [3H]-AMPA addition and measured after 40 mins by scint...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]-KA from rat GluK2More data for this Ligand-Target Pair