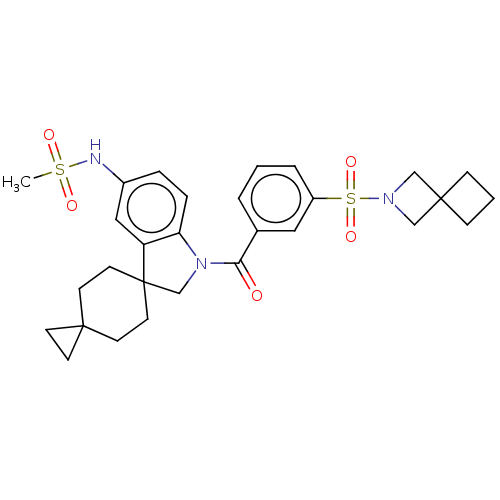
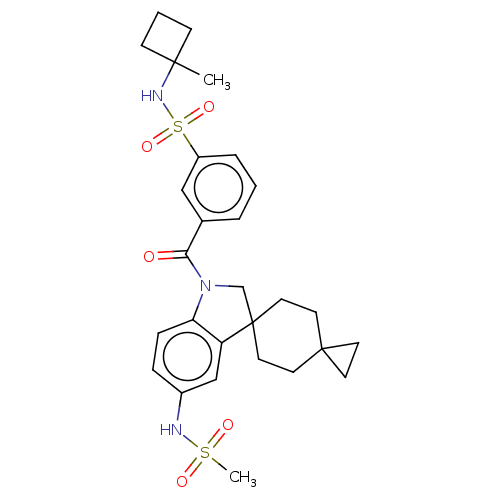
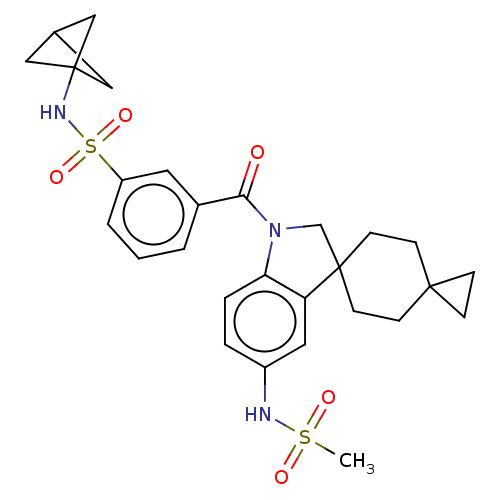
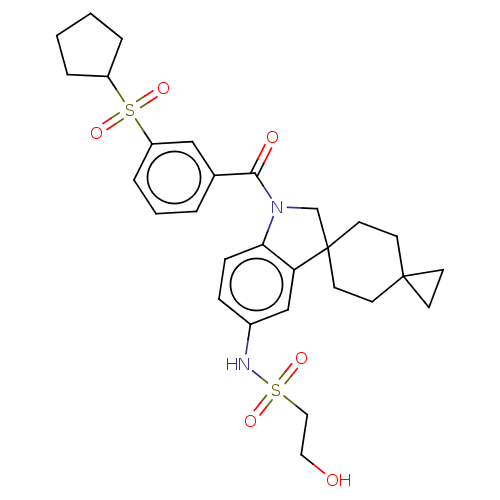
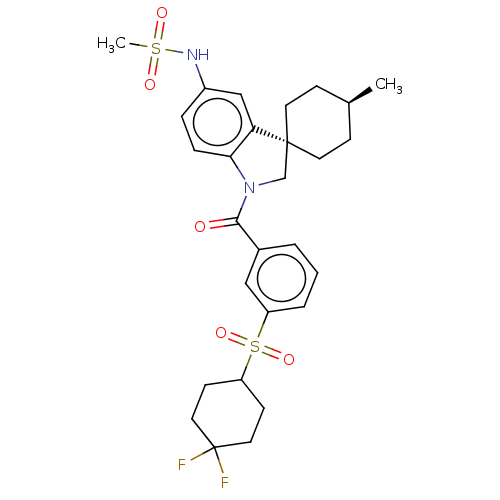
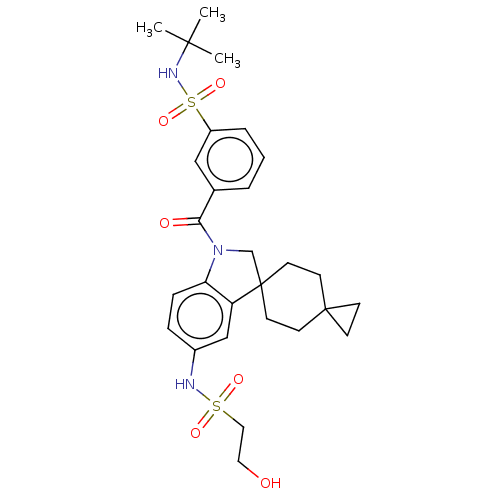
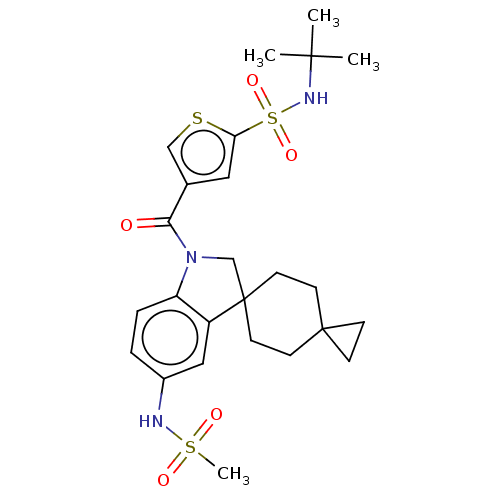
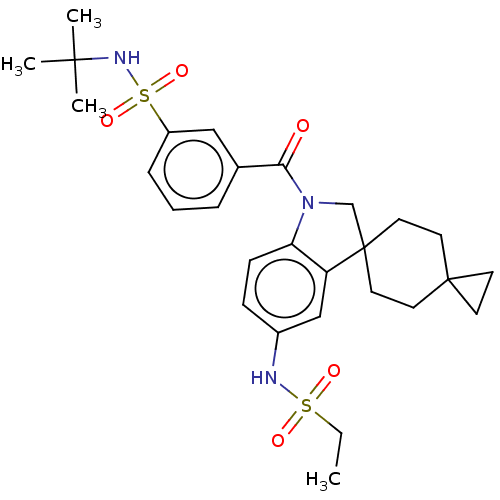
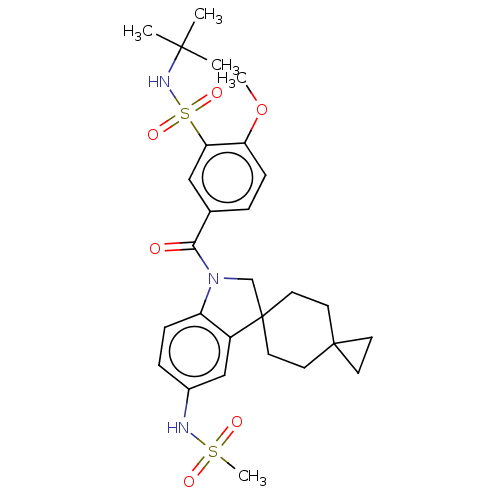
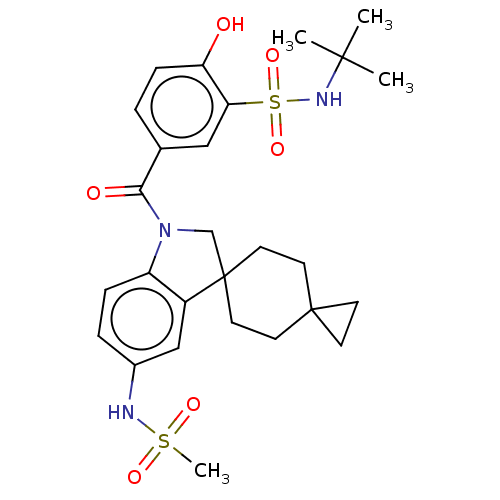
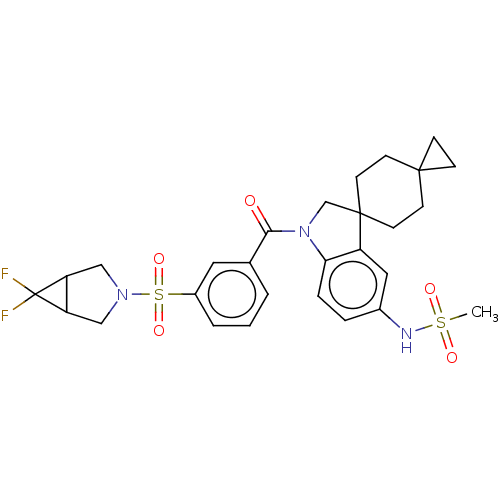
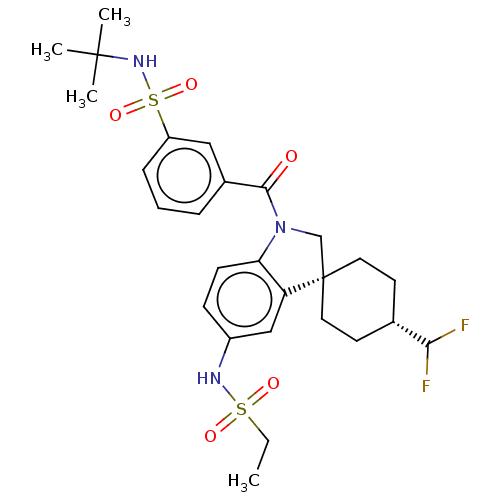
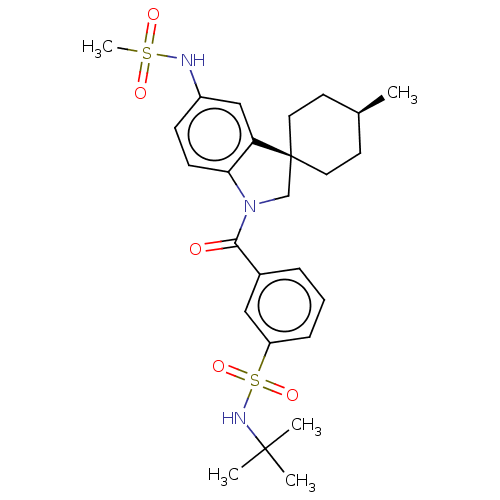
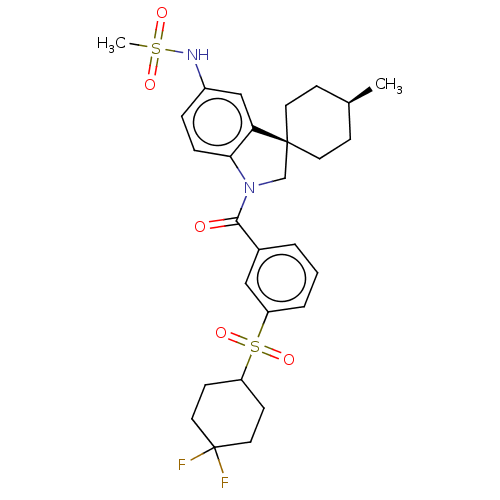
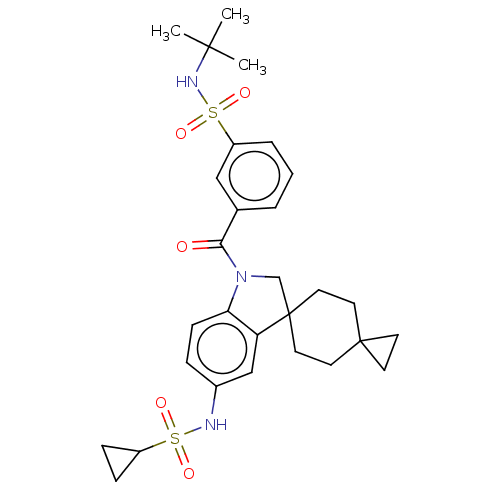
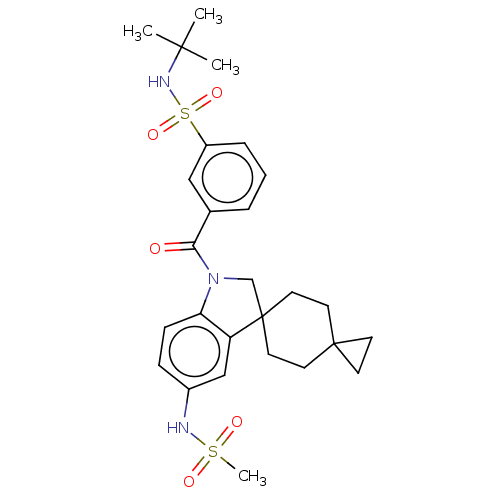
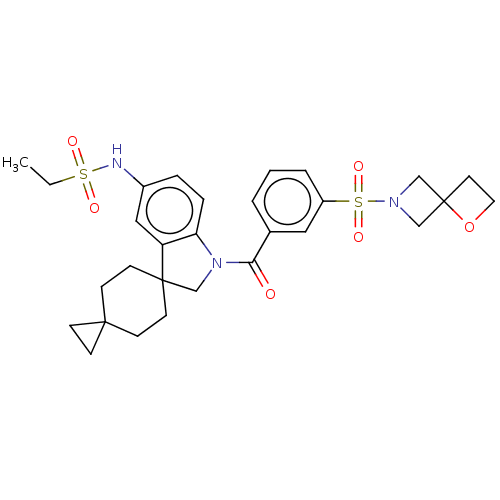
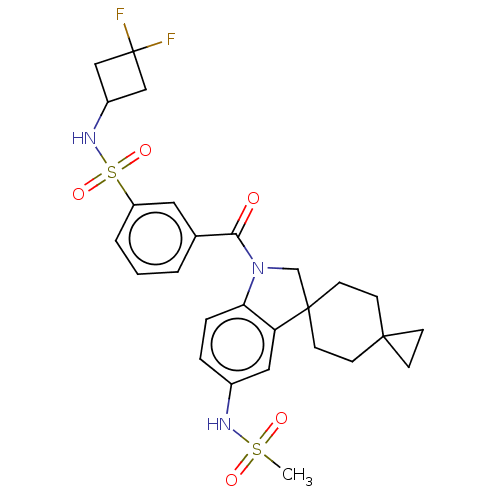
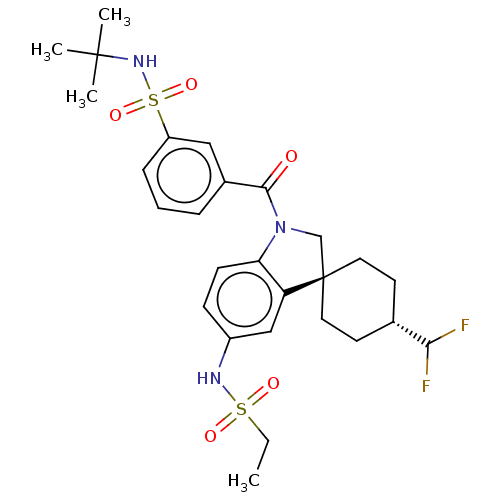
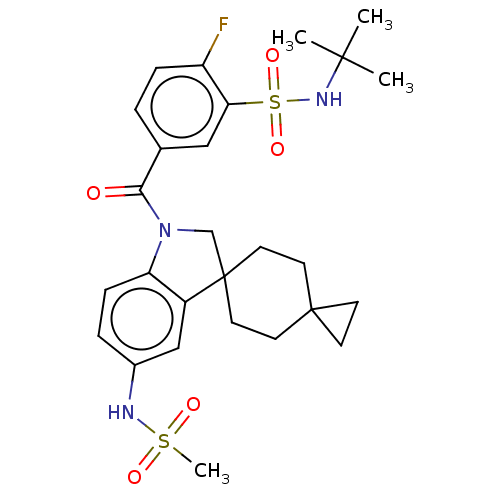
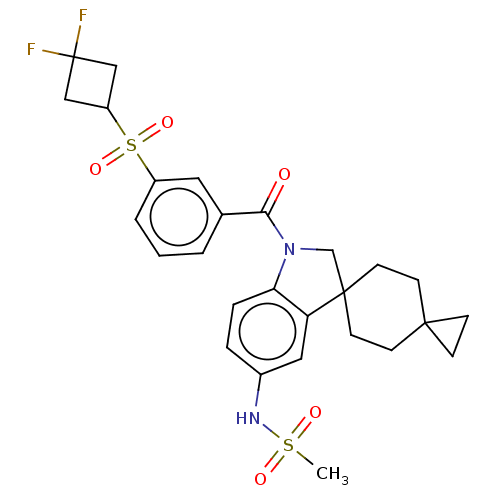
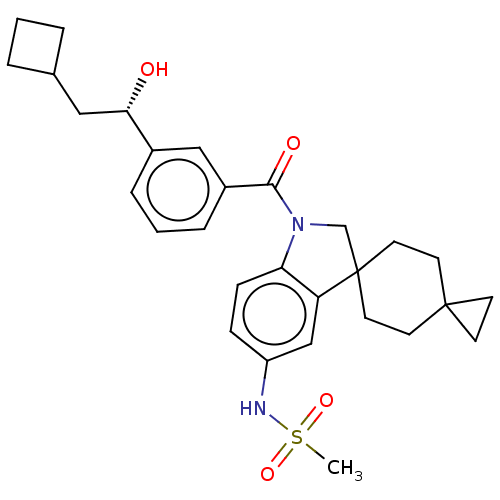
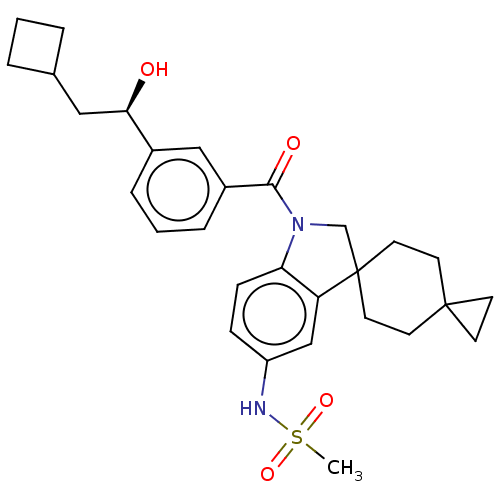
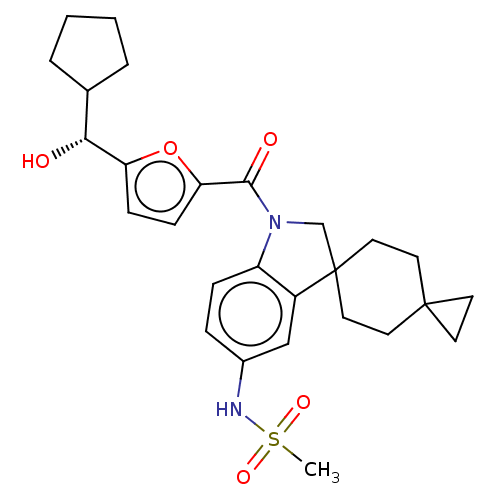
Report error Found 277 Enz. Inhib. hit(s) with all data for entry = 12324
Affinity DataKi: 0.120nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
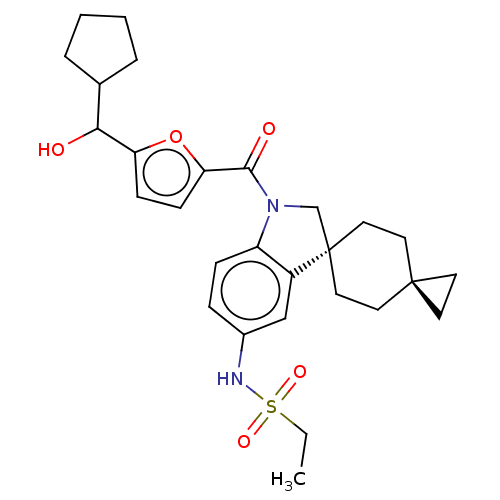
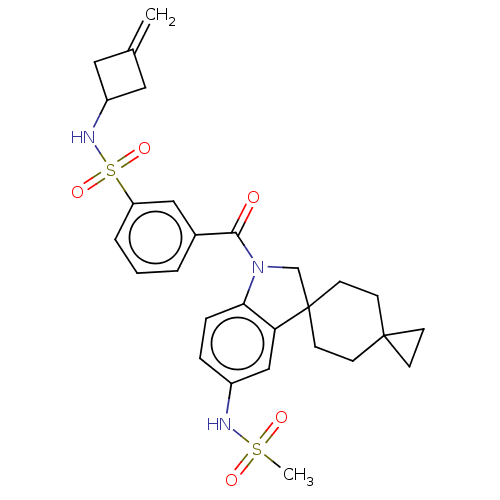
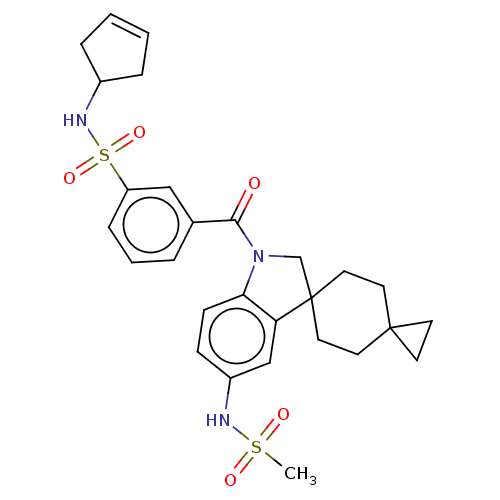
Ligand InfoSimilars
Affinity DataKi: 0.260nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
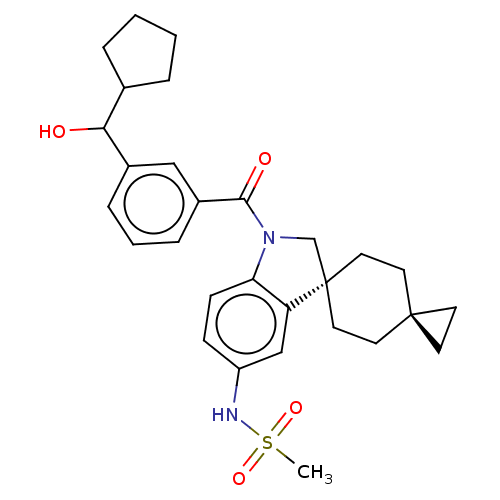
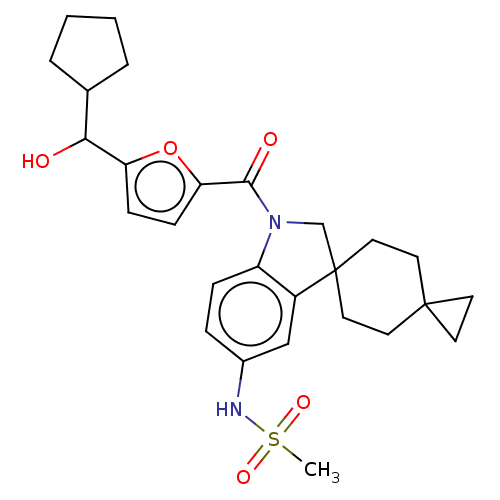
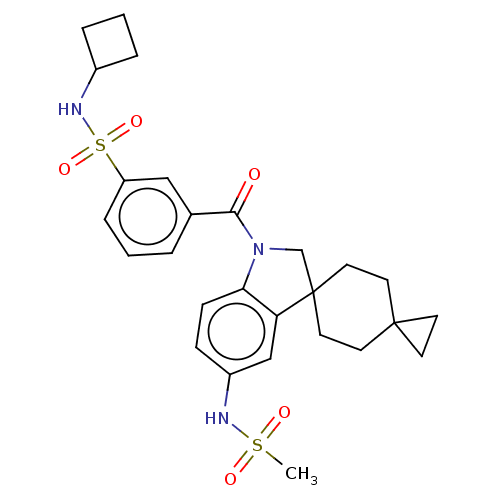
Ligand InfoSimilars
Affinity DataKi: 0.260nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
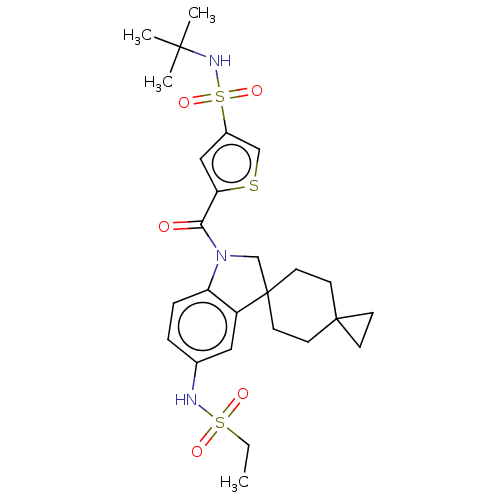
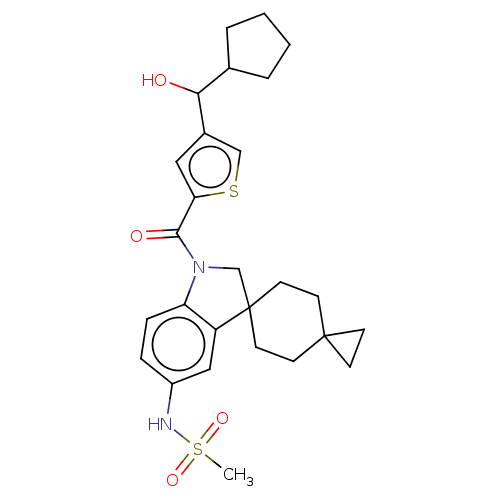
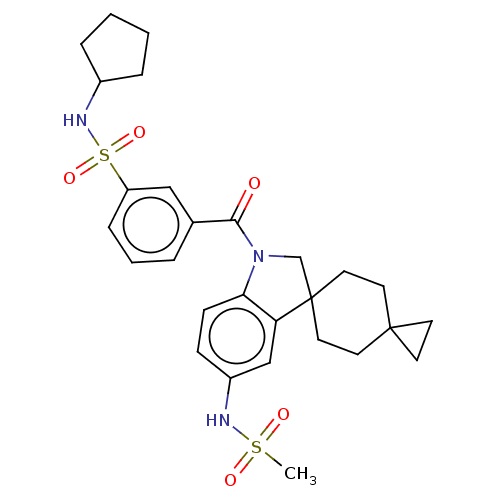
Ligand InfoSimilars
Affinity DataKi: 0.290nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
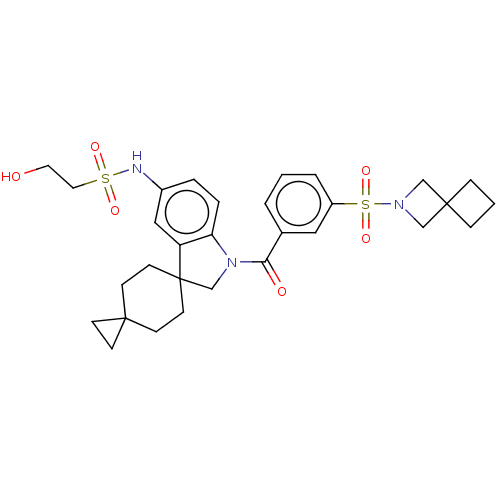
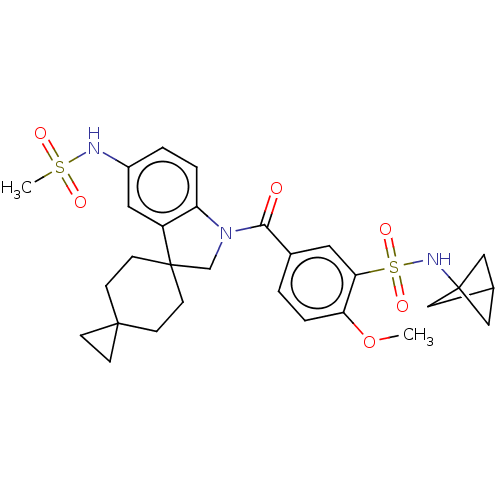
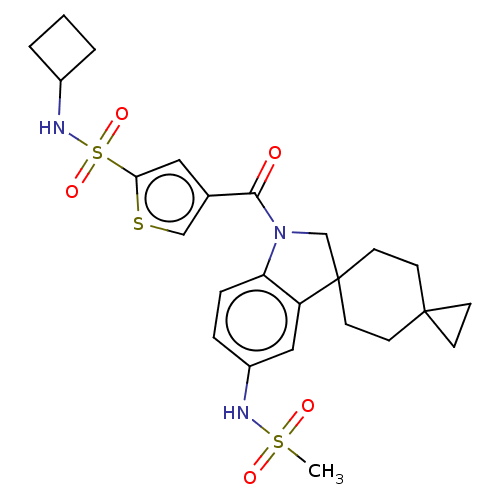
Ligand InfoSimilars
Affinity DataKi: 0.290nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.290nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.310nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.340nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.380nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.380nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.440nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.470nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.5nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.540nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.540nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.550nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.570nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.640nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.770nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.830nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 0.960nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 1.40nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 1.40nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 1.80nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 1.90nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 2nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 3nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.10nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.30nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 3.30nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.5nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 3.5nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.70nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.70nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 3.90nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 3.90nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 4nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 4nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 4.20nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 4.60nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 4.80nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 4.80nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.10nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.20nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.5nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.5nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.60nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 5.80nMAssay Description:Table 9: Test compounds were plated in a 3× dilution scheme in a 384-well plate. Assay buffer: 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataKi: 5.80nMAssay Description:Table 10: Compound binding kinetics parameters (kon and koff) were determined by the method of global progress curve analysis (GPCA). KIF18A (0.25 nM...More data for this Ligand-Target Pair
Ligand InfoSimilars