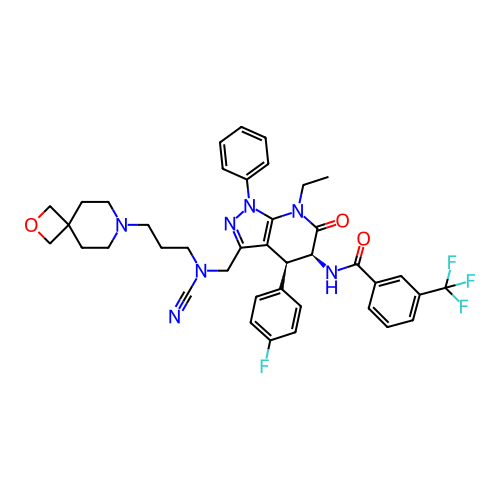
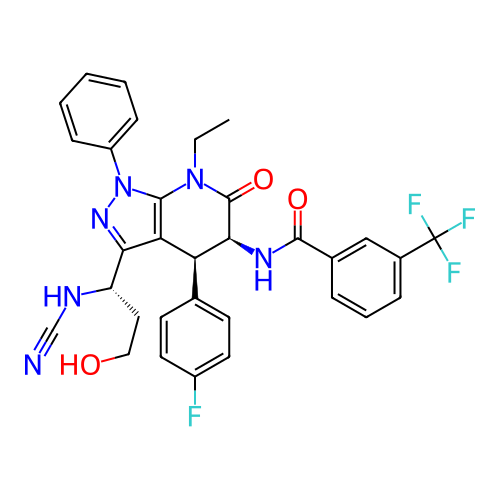
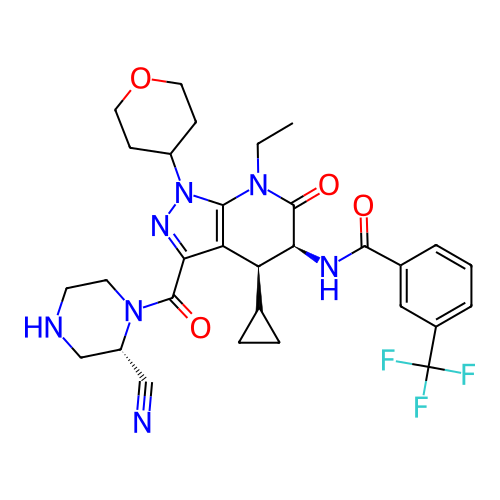
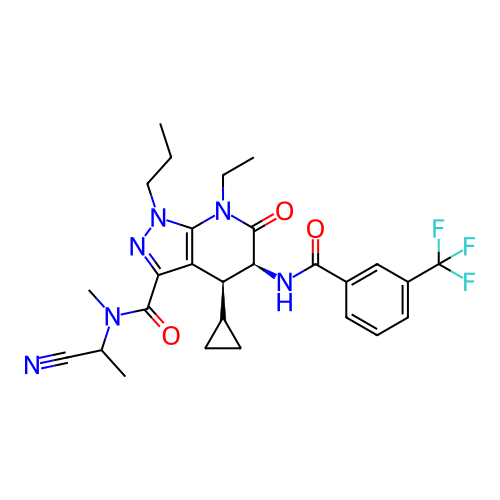
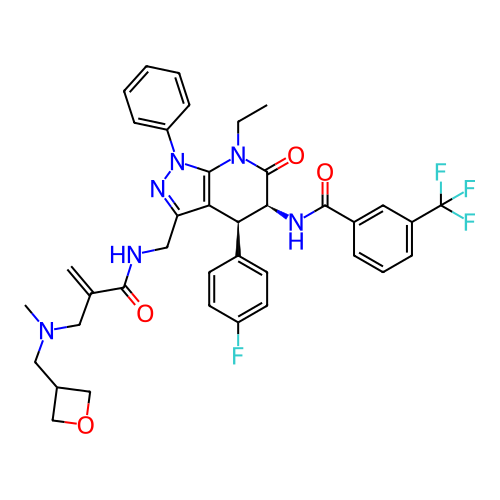
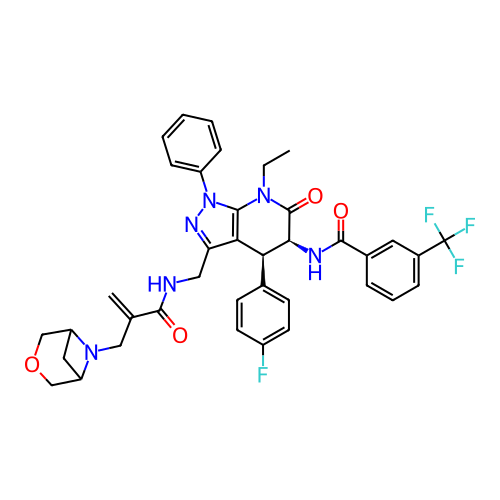
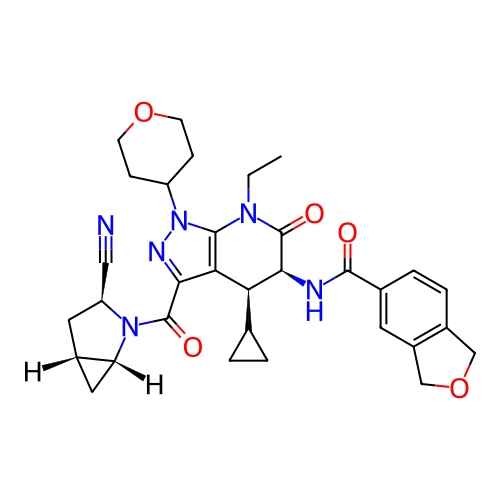
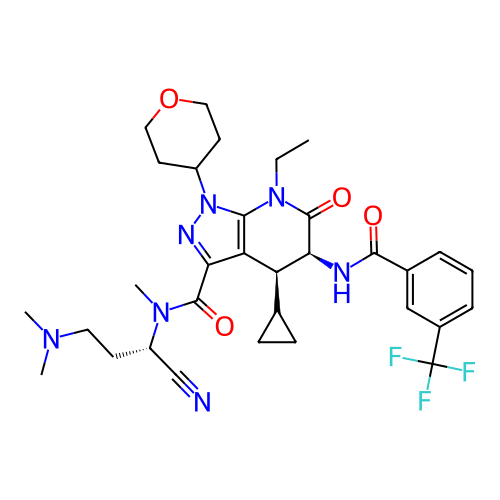
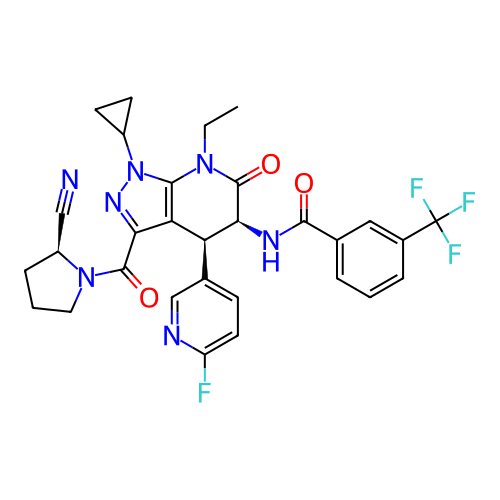
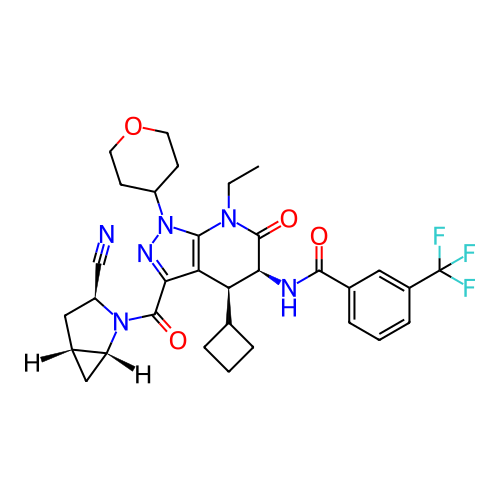
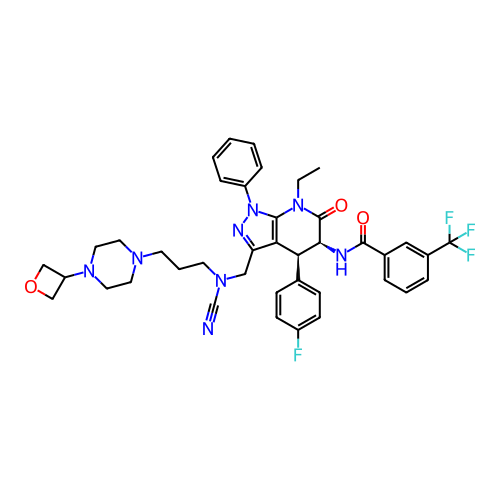
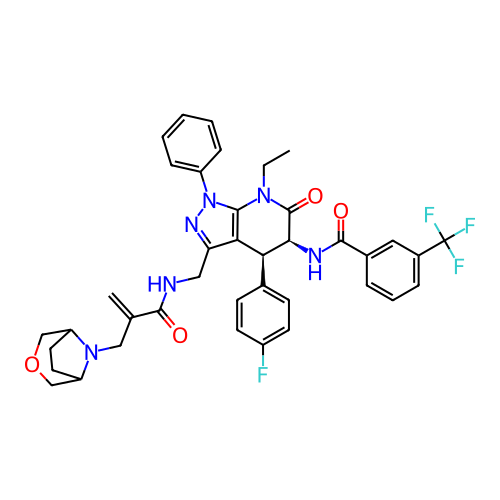
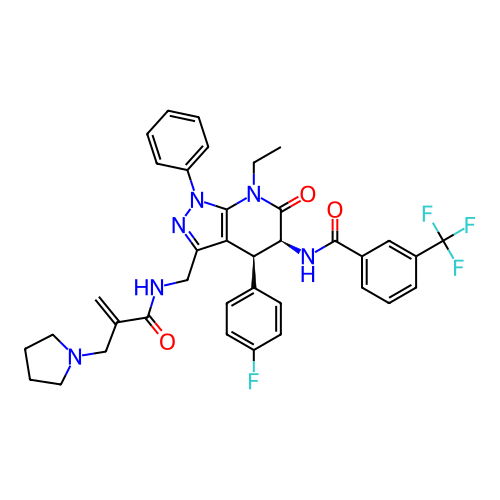
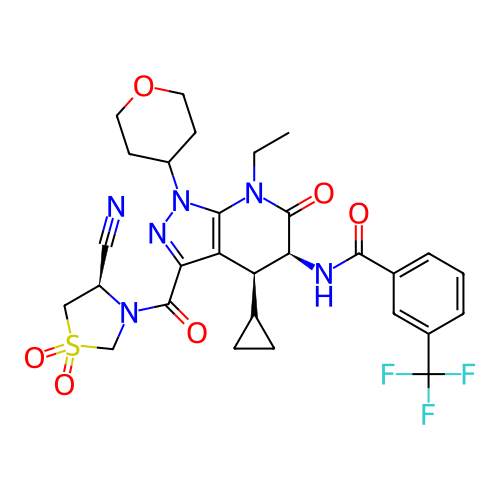
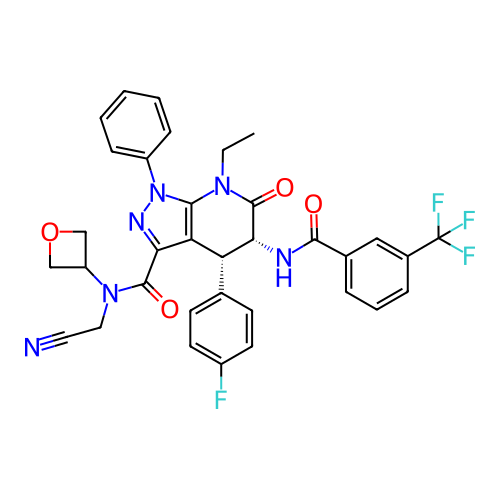
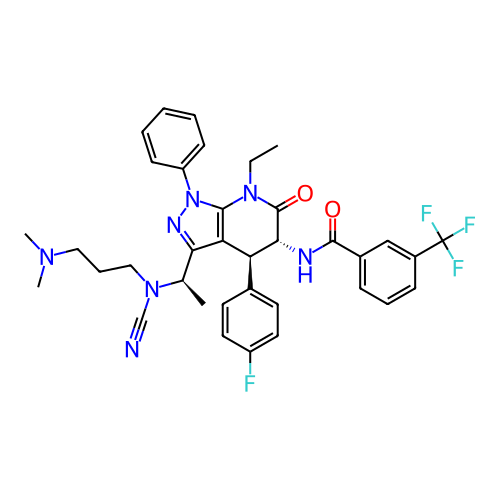
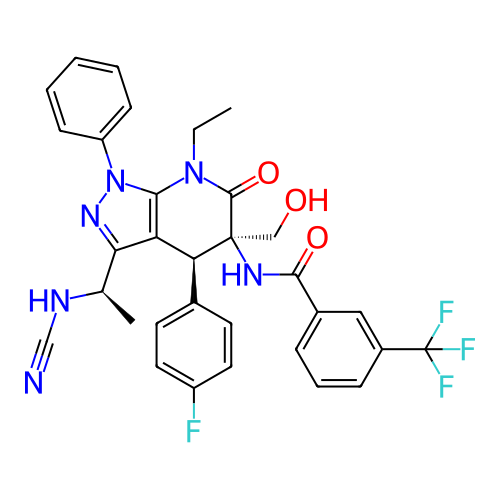
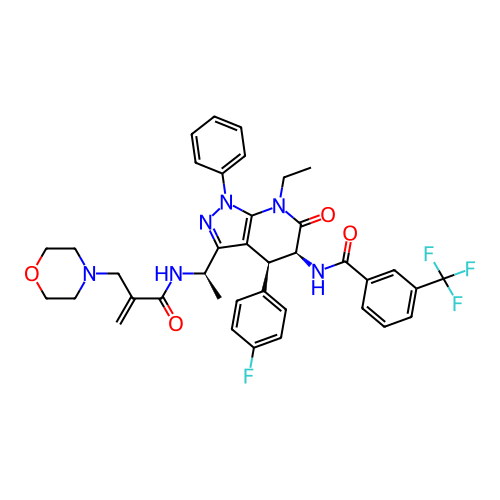
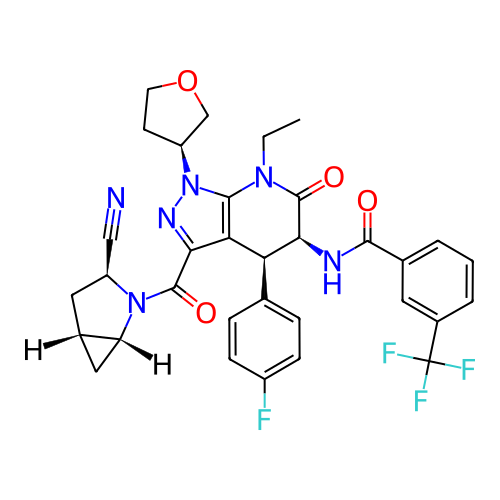
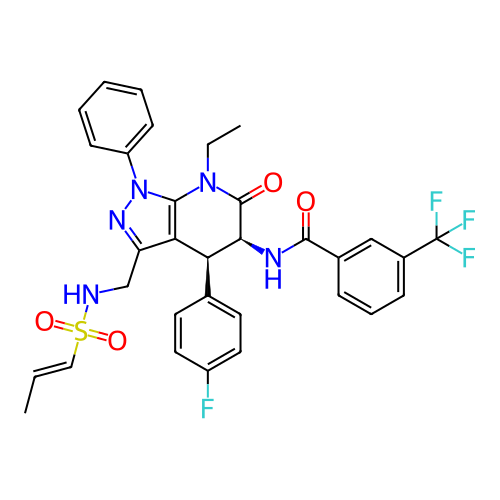
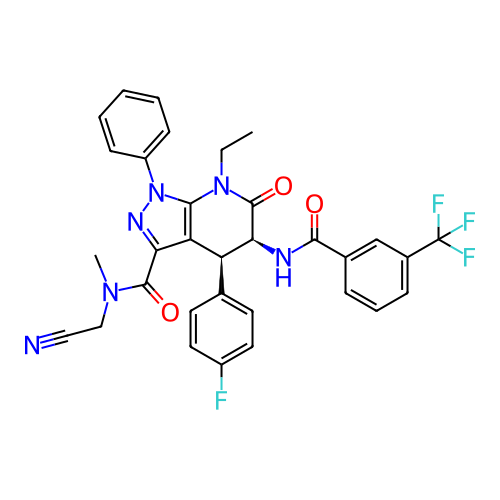
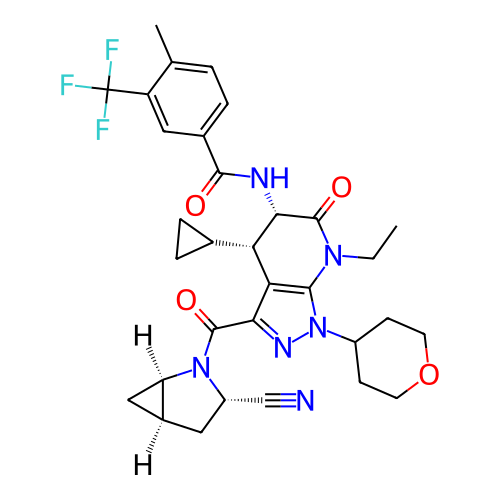
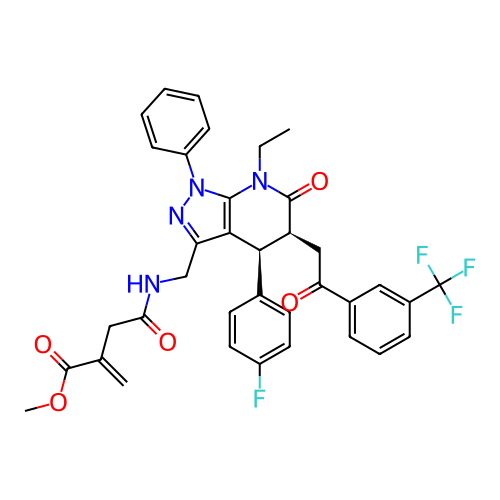
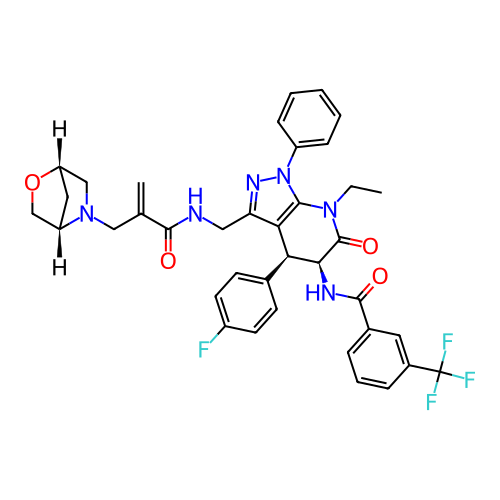
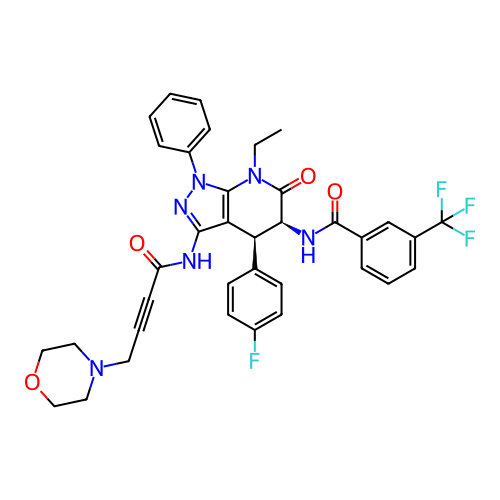
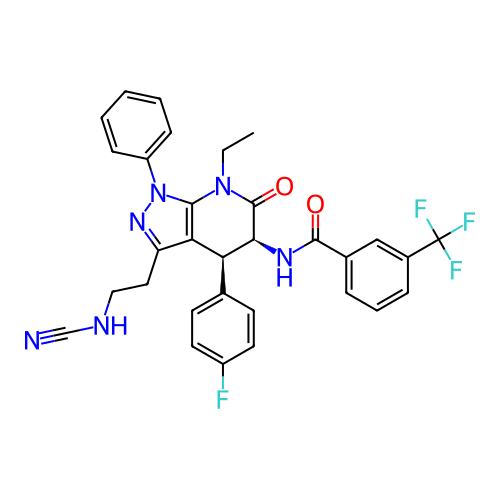
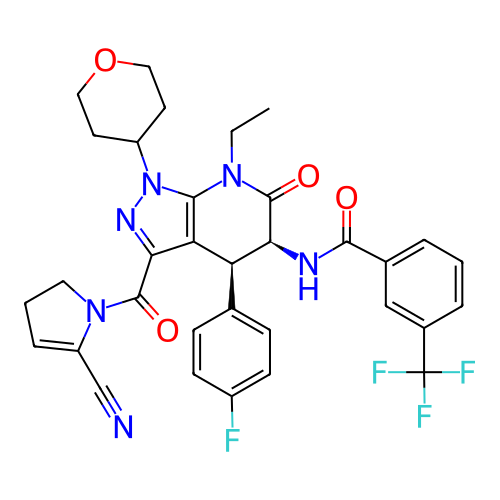
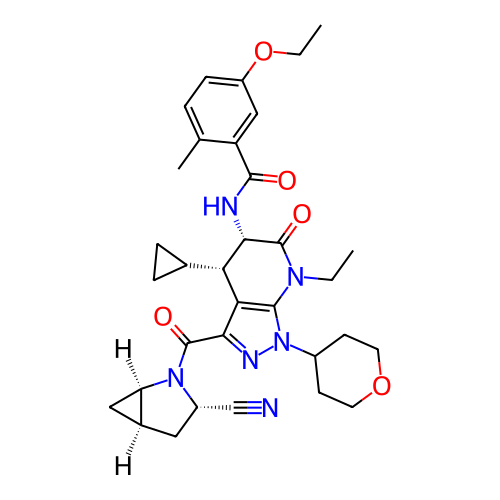
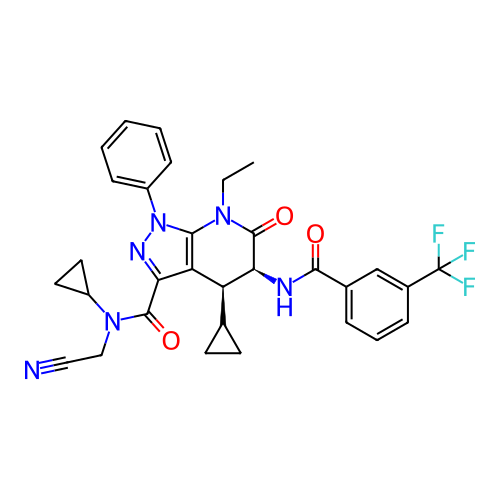
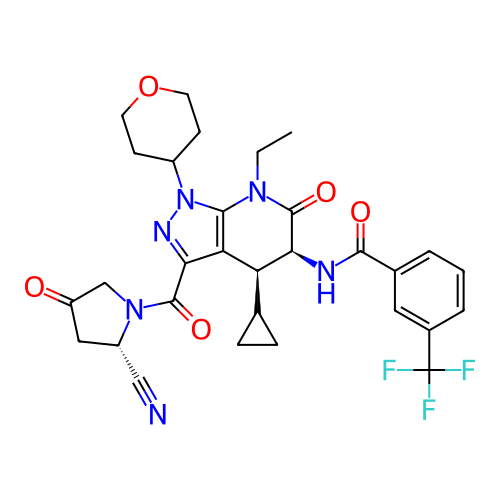
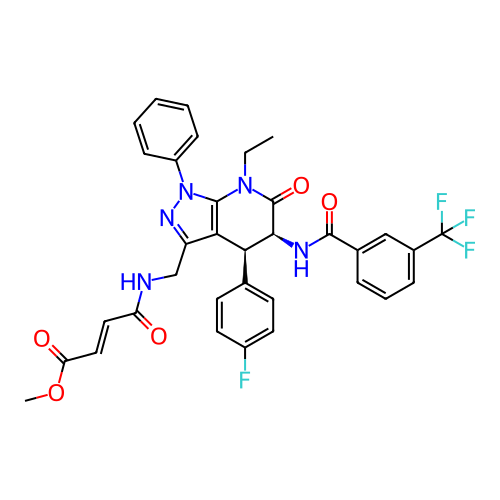
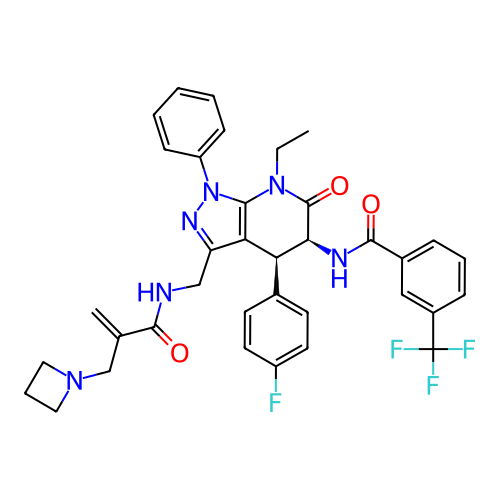
Report error Found 1174 Enz. Inhib. hit(s) with all data for entry = 12937
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:The TR-FRET assay was designed following the Scott et al. protocol (Scott et al., Nat Chem Biol. 2017 August; 13(8): 850-857. Doi:10.1038/nchembio.23...More data for this Ligand-Target Pair