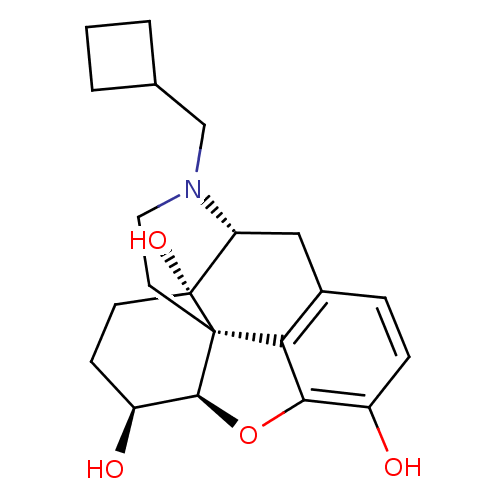
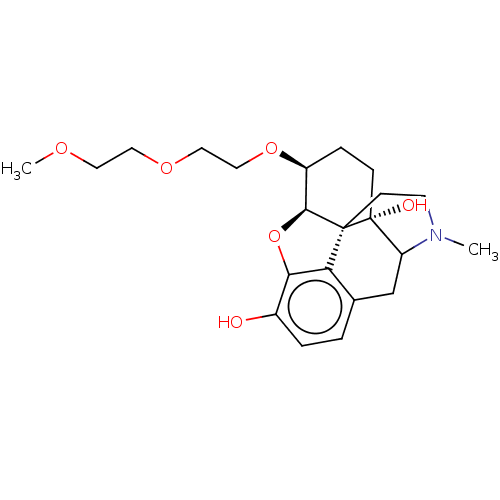
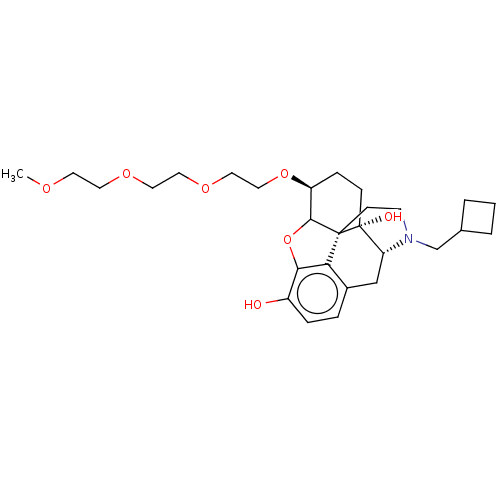
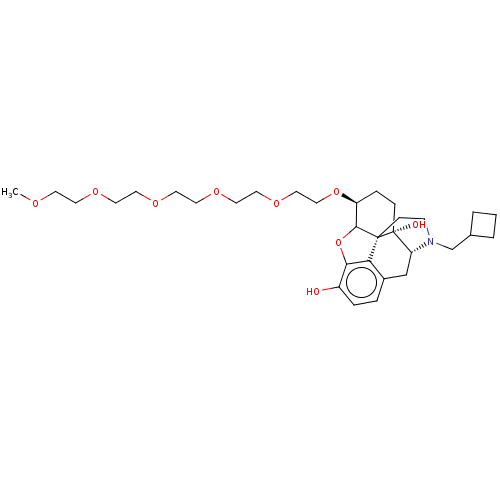
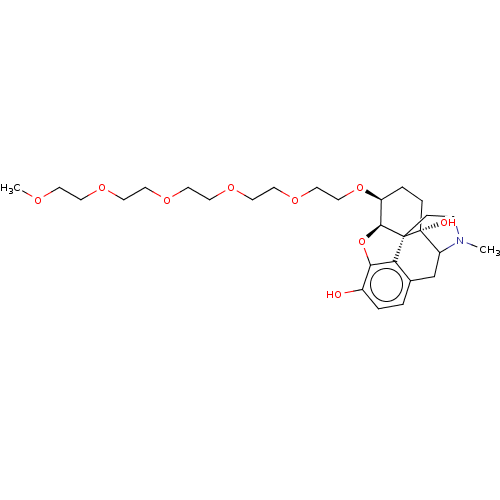
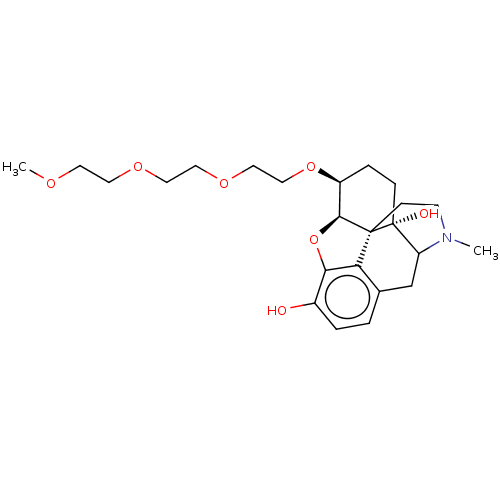
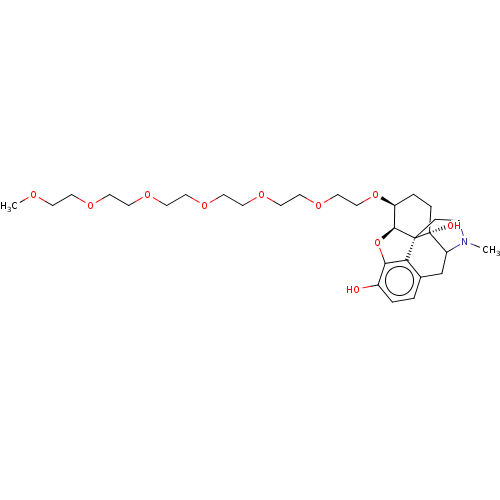
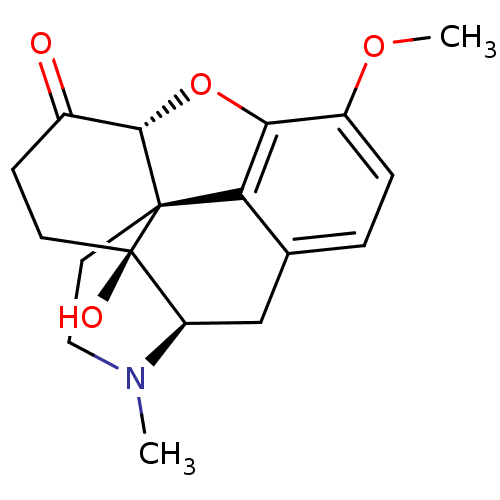
Report error Found 124 Enz. Inhib. hit(s) with all data for entry = 7646
Affinity DataKi: 8.44nM ΔG°: -46.1kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 14.3nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 15.7nM ΔG°: -44.5kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 17.5nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 22.0nM ΔG°: -43.7kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 23.1nM ΔG°: -43.6kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataEC50: 25.1nMpH: 7.4 T: 2°CAssay Description:Test compound and/or vehicle was preincubated with the cell membranes and 3 uM GDP in modified HEPES buffer (pH 7.4) for 20 minutes, followed by addi...More data for this Ligand-Target Pair
Affinity DataEC50: 26.8nMpH: 7.4 T: 2°CAssay Description:Test compound and/or vehicle was preincubated with the cell membranes and 3 uM GDP in modified HEPES buffer (pH 7.4) for 20 minutes, followed by addi...More data for this Ligand-Target Pair
Affinity DataKi: 27.4nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 28.5nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 29.9nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 35.1nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 35.6nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 39.4nM ΔG°: -42.3kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 44.3nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 50.7nM ΔG°: -41.6kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 56.9nM ΔG°: -41.4kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 73.0nM ΔG°: -40.7kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 77.9nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 79.5nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 85nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataEC50: 93.3nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 111nM ΔG°: -39.7kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 118nM ΔG°: -39.5kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 122nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 128nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 130nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 133nM ΔG°: -39.2kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 148nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 149nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 157nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 158nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 163nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 164nMpH: 7.4 T: 2°CAssay Description:Test compound and/or vehicle was preincubated with the cell membranes and 3 uM GDP in modified HEPES buffer (pH 7.4) for 20 minutes, followed by addi...More data for this Ligand-Target Pair
Affinity DataKi: 168nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 186nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 218nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 247nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataEC50: 270nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 282nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataIC50: 284nMpH: 7.4 T: 2°CAssay Description:Test compound and/or vehicle was preincubated with the cell membranes and 3 uM GDP in modified HEPES buffer (pH 7.4) for 20 minutes, followed by addi...More data for this Ligand-Target Pair
Affinity DataKi: 314nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 319nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataKi: 346nMAssay Description:Specific binding is determined by subtraction of the cpm bound in the presence of 50-100× excess of cold ligand. Binding data assays were analyzed us...More data for this Ligand-Target Pair
Affinity DataIC50: 398nMpH: 7.4 T: 2°CAssay Description:Test compound and/or vehicle was preincubated with the cell membranes and 3 uM GDP in modified HEPES buffer (pH 7.4) for 20 minutes, followed by addi...More data for this Ligand-Target Pair
Affinity DataKi: 404nM ΔG°: -36.5kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataEC50: 415nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair
Affinity DataKi: 439nM ΔG°: -36.3kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataKi: 445nM ΔG°: -36.3kJ/moleT: 2°CAssay Description:Briefly, serial dilutions of the test compounds were placed in a 96-well plate to which were added SPA beads, membrane and radioligand. The assay con...More data for this Ligand-Target Pair
Affinity DataEC50: 478nMT: 2°CAssay Description:Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xan...More data for this Ligand-Target Pair