Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Histone deacetylase 4 [653-1084,H976Y]
Ligand
BDBM19410
Substrate
BDBM25143
Meas. Tech.
HDAC Activity Assay
Temperature
298.15±n/a K
IC50
>10000±n/a nM
Comments
extracted
Citation
 Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem 51:2350-3 (2008) [PubMed] Article
Jones, P; Altamura, S; De Francesco, R; Paz, OG; Kinzel, O; Mesiti, G; Monteagudo, E; Pescatore, G; Rowley, M; Verdirame, M; Steinkühler, C A novel series of potent and selective ketone histone deacetylase inhibitors with antitumor activity in vivo. J Med Chem 51:2350-3 (2008) [PubMed] Article More Info.:
Target
Name:
Histone deacetylase 4 [653-1084,H976Y]
Synonyms:
HDAC4 | HDAC4_HUMAN | Histone Deacetylase 4 (HDAC4) Mutant (H976Y) | Histone deacetylase 4 gain of function (GOF) mutant | KIAA0288
Type:
Catalytic domain
Mol. Mass.:
46476.72
Organism:
Homo sapiens (Human)
Description:
His-tagged HDAC4 GOF(gain of function, H976Y) catalytic domains (T653-L1084) was expressed in E.coli and purified by Nickel-Chelation affinity chromatography and anion exchange (MonoQ) chromatography.
Residue:
432
Sequence:
TTGLVYDTLMLKHQCTCGSSSSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQTVHSEAHTLLYGTNPLNRQKLDSKKLLGSLASVFVRLPCGGVGVDSDTIWNEVHSAGAARLAVGCVVELVFKVATGELKNGFAVVRPPGHHAEESTPMGFCYFNSVAVAAKLLQQRLSVSKILIVDWDVHHGNGTQQAFYSDPSVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLDPPMGDAEYLAAFRTVVMPIASEFAPDVVLVSSGFDAVEGHPTPLGGYNLSARCFGYLTKQLMGLAGGRIVLALEGGYDLTAICDASEACVSALLGNELDPLPEKVLQQRPNANAVRSMEKVMEIHSKYWRCLQRTTSTAGRSLIEAQTCENEEAETVTAMASLSVGVKPAEKRPDEEPMEEEPPL
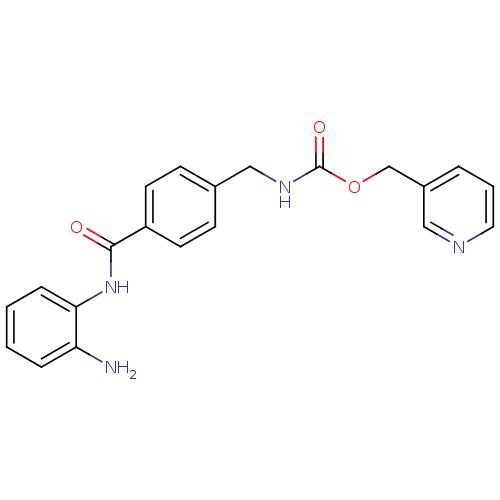
Inhibitor
Name:
BDBM19410
Synonyms:
CHEMBL27759 | MS-275 | US11377423, MS-275 | US11672788, Compound Entinostat | US9265734, MS-275 | US9796664, Compound MS-275 | benzamide-type inhibitor, 3 | pyridin-3-ylmethyl N-({4-[(2-aminophenyl)carbamoyl]phenyl}methyl)carbamate
Type:
Small organic molecule
Emp. Form.:
C21H20N4O3
Mol. Mass.:
376.4085
SMILES:
Nc1ccccc1NC(=O)c1ccc(CNC(=O)OCc2cccnc2)cc1