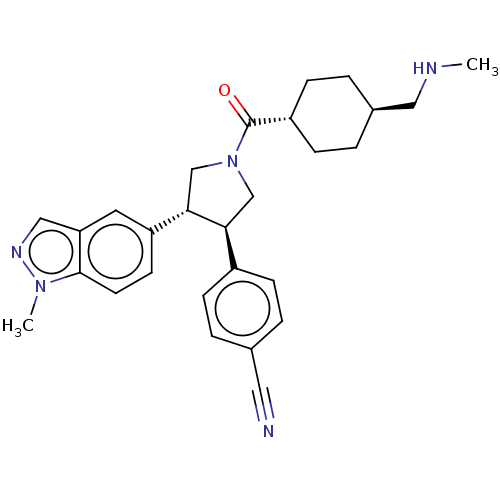
Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Lysine-specific histone demethylase 1A
Ligand
BDBM50262072
Substrate
n/a
Meas. Tech.
ChEMBL_1696558 (CHEMBL4047448)
Kd
540±n/a nM
Citation
 Mould, DP; Bremberg, U; Jordan, AM; Geitmann, M; McGonagle, AE; Somervaille, TCP; Spencer, GJ; Ogilvie, DJ Development and evaluation of 4-(pyrrolidin-3-yl)benzonitrile derivatives as inhibitors of lysine specific demethylase 1. Bioorg Med Chem Lett 27:4755-4759 (2017) [PubMed] Article
Mould, DP; Bremberg, U; Jordan, AM; Geitmann, M; McGonagle, AE; Somervaille, TCP; Spencer, GJ; Ogilvie, DJ Development and evaluation of 4-(pyrrolidin-3-yl)benzonitrile derivatives as inhibitors of lysine specific demethylase 1. Bioorg Med Chem Lett 27:4755-4759 (2017) [PubMed] Article More Info.:
Target
Name:
Lysine-specific histone demethylase 1A
Synonyms:
AOF2 | BRAF35-HDAC complex protein BHC110 | Flavin-containing amine oxidase domain-containing protein 2 | KDM1 | KDM1A | KDM1A_HUMAN | KIAA0601 | LSD1 | Lysine-specific demethylase 1 (LSD1) | Lysine-specific histone demethylase 1 | Lysine-specific histone demethylase 1 (LSD1)
Type:
Enzyme
Mol. Mass.:
92901.01
Organism:
Homo sapiens (Human)
Description:
O60341
Residue:
852
Sequence:
MLSGKKAAAAAAAAAAAATGTEAGPGTAGGSENGSEVAAQPAGLSGPAEVGPGAVGERTPRKKEPPRASPPGGLAEPPGSAGPQAGPTVVPGSATPMETGIAETPEGRRTSRRKRAKVEYREMDESLANLSEDEYYSEEERNAKAEKEKKLPPPPPQAPPEEENESEPEEPSGVEGAAFQSRLPHDRMTSQEAACFPDIISGPQQTQKVFLFIRNRTLQLWLDNPKIQLTFEATLQQLEAPYNSDTVLVHRVHSYLERHGLINFGIYKRIKPLPTKKTGKVIIIGSGVSGLAAARQLQSFGMDVTLLEARDRVGGRVATFRKGNYVADLGAMVVTGLGGNPMAVVSKQVNMELAKIKQKCPLYEANGQAVPKEKDEMVEQEFNRLLEATSYLSHQLDFNVLNNKPVSLGQALEVVIQLQEKHVKDEQIEHWKKIVKTQEELKELLNKMVNLKEKIKELHQQYKEASEVKPPRDITAEFLVKSKHRDLTALCKEYDELAETQGKLEEKLQELEANPPSDVYLSSRDRQILDWHFANLEFANATPLSTLSLKHWDQDDDFEFTGSHLTVRNGYSCVPVALAEGLDIKLNTAVRQVRYTASGCEVIAVNTRSTSQTFIYKCDAVLCTLPLGVLKQQPPAVQFVPPLPEWKTSAVQRMGFGNLNKVVLCFDRVFWDPSVNLFGHVGSTTASRGELFLFWNLYKAPILLALVAGEAAGIMENISDDVIVGRCLAILKGIFGSSAVPQPKETVVSRWRADPWARGSYSYVAAGSSGNDYDLMAQPITPGPSIPGAPQPIPRLFFAGEHTIRNYPATVHGALLSGLREAGRIADQFLGAMYTLPRQATPGVPAQQSPSM
Inhibitor
Name:
BDBM50262072
Synonyms:
CHEMBL4079770
Type:
Small organic molecule
Emp. Form.:
C28H33N5O
Mol. Mass.:
455.5945
SMILES:
CNC[C@H]1CC[C@@H](CC1)C(=O)N1C[C@H]([C@@H](C1)c1ccc2n(C)ncc2c1)c1ccc(cc1)C#N |r,wU:6.9,13.29,wD:14.17,3.2,(68.45,-37.02,;69.8,-36.27,;69.82,-34.74,;68.49,-33.95,;68.51,-32.41,;67.19,-31.62,;65.85,-32.38,;65.83,-33.92,;67.15,-34.7,;64.53,-31.6,;63.18,-32.35,;64.55,-30.06,;63.32,-29.12,;63.8,-27.66,;65.35,-27.68,;65.81,-29.16,;66.28,-26.45,;65.66,-25.03,;66.58,-23.8,;68.12,-23.98,;69.28,-22.96,;69.13,-21.43,;70.61,-23.74,;70.27,-25.25,;68.73,-25.4,;67.81,-26.63,;62.92,-26.41,;63.55,-25,;62.67,-23.75,;61.13,-23.89,;60.48,-25.29,;61.38,-26.55,;60.25,-22.63,;59.35,-21.36,)|