Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Lysine-specific demethylase 4B
Ligand
BDBM191600
Substrate
n/a
Meas. Tech.
ChEMBL_1771188 (CHEMBL4223300)
IC50
170±n/a nM
Citation
 Nie, Z; Shi, L; Lai, C; O'Connell, SM; Xu, J; Stansfield, RK; Hosfield, DJ; Veal, JM; Stafford, JA Structure-based design and discovery of potent and selective KDM5 inhibitors. Bioorg Med Chem Lett 28:1490-1494 (2018) [PubMed] Article
Nie, Z; Shi, L; Lai, C; O'Connell, SM; Xu, J; Stansfield, RK; Hosfield, DJ; Veal, JM; Stafford, JA Structure-based design and discovery of potent and selective KDM5 inhibitors. Bioorg Med Chem Lett 28:1490-1494 (2018) [PubMed] Article More Info.:
Target
Name:
Lysine-specific demethylase 4B
Synonyms:
JHDM3B | JMJD2B | JmjC domain-containing histone demethylation protein 3B | Jumonji domain-containing protein 2B | KDM4B | KDM4B_HUMAN | KIAA0876
Type:
PROTEIN
Mol. Mass.:
121904.33
Organism:
Homo sapiens (Human)
Description:
ChEMBL_109446
Residue:
1096
Sequence:
MGSEDHGAQNPSCKIMTFRPTMEEFKDFNKYVAYIESQGAHRAGLAKIIPPKEWKPRQTYDDIDDVVIPAPIQQVVTGQSGLFTQYNIQKKAMTVGEYRRLANSEKYCTPRHQDFDDLERKYWKNLTFVSPIYGADISGSLYDDDVAQWNIGSLRTILDMVERECGTIIEGVNTPYLYFGMWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAIGFFPGSSQGCDAFLRHKMTLISPIILKKYGIPFSRITQEAGEFMITFPYGYHAGFNHGFNCAESTNFATLRWIDYGKVATQCTCRKDMVKISMDVFVRILQPERYELWKQGKDLTVLDHTRPTALTSPELSSWSASRASLKAKLLRRSHRKRSQPKKPKPEDPKFPGEGTAGAALLEEAGGSVKEEAGPEVDPEEEEEEPQPLPHGREAEGAEEDGRGKLRPTKAKSERKKKSFGLLPPQLPPPPAHFPSEEALWLPSPLEPPVLGPGPAAMEESPLPAPLNVVPPEVPSEELEAKPRPIIPMLYVVPRPGKAAFNQEHVSCQQAFEHFAQKGPTWKEPVSPMELTGPEDGAASSGAGRMETKARAGEGQAPSTFSKLKMEIKKSRRHPLGRPPTRSPLSVVKQEASSDEEASPFSGEEDVSDPDALRPLLSLQWKNRAASFQAERKFNAAAARTEPYCAICTLFYPYCQALQTEKEAPIASLGKGCPATLPSKSRQKTRPLIPEMCFTSGGENTEPLPANSYIGDDGTSPLIACGKCCLQVHASCYGIRPELVNEGWTCSRCAAHAWTAECCLCNLRGGALQMTTDRRWIHVICAIAVPEARFLNVIERHPVDISAIPEQRWKLKCVYCRKRMKKVSGACIQCSYEHCSTSFHVTCAHAAGVLMEPDDWPYVVSITCLKHKSGGHAVQLLRAVSLGQVVITKNRNGLYYRCRVIGAASQTCYEVNFDDGSYSDNLYPESITSRDCVQLGPPSEGELVELRWTDGNLYKAKFISSVTSHIYQVEFEDGSQLTVKRGDIFTLEEELPKRVRSRLSLSTGAPQEPAFSGEEAKAAKRPRVGTPLATEDSGRSQDYVAFVESLLQVQGRPGAPF
Inhibitor
Name:
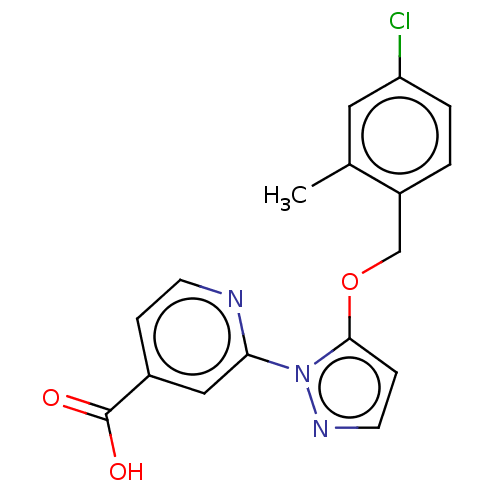
BDBM191600
Synonyms:
2-(5-((4-chloro-2-methylbenzyl)oxy)-1Hpyrazol-1-yl)isonicotinic acid (N19) | US10173996, Example 89 | US9604961, Example 89 | US9714230, 89 | US9908865, Example 89
Type:
Small organic molecule
Emp. Form.:
C17H14ClN3O3
Mol. Mass.:
343.764
SMILES:
Cc1cc(Cl)ccc1COc1ccnn1-c1cc(ccn1)C(O)=O