Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Gap junction beta-2 protein
Ligand
BDBM50565239
Substrate
n/a
Meas. Tech.
ChEMBL_2091062 (CHEMBL4772325)
IC50
12700±n/a nM
Citation
 Subedi, YP; Kjellgren, A; Roberts, P; Montgomery, H; Thackeray, N; Fiori, MC; Altenberg, GA; Chang, CT Amphiphilic aminoglycosides with increased selectivity for inhibition of connexin 43 (Cx43) hemichannels. Eur J Med Chem 203:0 (2020) [PubMed] Article
Subedi, YP; Kjellgren, A; Roberts, P; Montgomery, H; Thackeray, N; Fiori, MC; Altenberg, GA; Chang, CT Amphiphilic aminoglycosides with increased selectivity for inhibition of connexin 43 (Cx43) hemichannels. Eur J Med Chem 203:0 (2020) [PubMed] Article More Info.:
Target
Name:
Gap junction beta-2 protein
Synonyms:
CXB2_HUMAN | Connexin-26 | Cx26 | GJB2 | Gap junction beta-2 protein
Type:
PROTEIN
Mol. Mass.:
26226.37
Organism:
Homo sapiens
Description:
ChEMBL_117983
Residue:
226
Sequence:
MDWGTLQTILGGVNKHSTSIGKIWLTVLFIFRIMILVVAAKEVWGDEQADFVCNTLQPGCKNVCYDHYFPISHIRLWALQLIFVSTPALLVAMHVAYRRHEKKRKFIKGEIKSEFKDIEEIKTQKVRIEGSLWWTYTSSIFFRVIFEAAFMYVFYVMYDGFSMQRLVKCNAWPCPNTVDCFVSRPTEKTVFTVFMIAVSGICILLNVTELCYLLIRYCSGKSKKPV
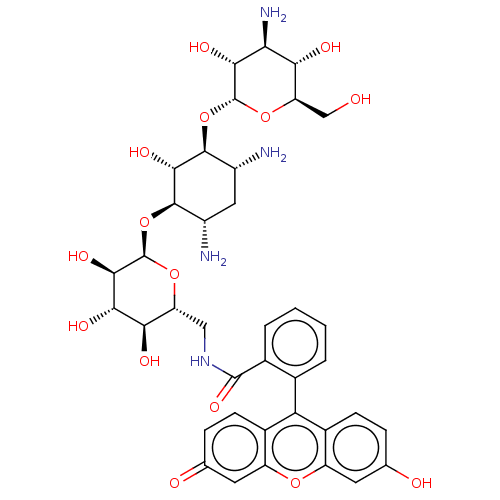
Inhibitor
Name:
BDBM50565239
Synonyms:
CHEMBL4785499
Type:
Small organic molecule
Emp. Form.:
C38H46N4O15
Mol. Mass.:
798.7896
SMILES:
[H][C@]1(O[C@H]2[C@H](N)C[C@H](N)[C@@H](O[C@@]3([H])O[C@H](CNC(=O)c4ccccc4-c4c5ccc(O)cc5oc5cc(=O)ccc45)[C@@H](O)[C@H](O)[C@H]3O)[C@@H]2O)O[C@H](CO)[C@@H](O)[C@H](N)[C@H]1O |r,wU:43.47,50.56,9.9,14.14,11.11,3.2,55.61,1.0,wD:53.59,45.50,4.4,41.45,47.53,57.64,7.7,(8.96,-8.44,;8.1,-7.17,;9.65,-7.17,;10.41,-5.83,;9.65,-4.51,;8.1,-4.51,;10.41,-3.17,;11.95,-3.17,;12.72,-1.83,;12.72,-4.51,;14.26,-4.51,;15.03,-5.83,;15.47,-4.35,;14.26,-7.17,;15.03,-8.5,;14.26,-9.84,;15.03,-11.17,;14.26,-12.51,;12.92,-11.73,;15.03,-13.84,;16.56,-13.83,;17.33,-15.16,;16.56,-16.5,;15.02,-16.5,;14.25,-15.17,;12.71,-15.16,;11.94,-16.49,;12.71,-17.82,;11.94,-19.16,;10.39,-19.16,;9.62,-20.49,;9.62,-17.82,;10.4,-16.49,;9.63,-15.15,;10.41,-13.82,;9.64,-12.48,;10.42,-11.15,;9.65,-9.82,;11.96,-11.16,;12.72,-12.49,;11.95,-13.83,;16.57,-8.5,;17.34,-9.84,;17.34,-7.17,;18.89,-7.17,;16.57,-5.83,;17.34,-4.51,;11.95,-5.83,;12.72,-7.17,;7.33,-5.83,;5.78,-5.83,;5.02,-4.51,;3.47,-4.51,;5.02,-7.17,;3.47,-7.17,;5.78,-8.5,;5.02,-9.84,;7.33,-8.5,;8.1,-9.84,)|