Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Protein mono-ADP-ribosyltransferase PARP3
Ligand
BDBM50579974
Substrate
n/a
Meas. Tech.
ChEMBL_2146113 (CHEMBL5030393)
IC50
>10000±n/a nM
Citation
 Leenders, RGG; Brinch, SA; Sowa, ST; Amundsen-Isaksen, E; Galera-Prat, A; Murthy, S; Aertssen, S; Smits, JN; Nieczypor, P; Damen, E; Wegert, A; Nazaré, M; Lehtiö, L; Waaler, J; Krauss, S Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II. J Med Chem 64:17936-17949 (2021) [PubMed] Article
Leenders, RGG; Brinch, SA; Sowa, ST; Amundsen-Isaksen, E; Galera-Prat, A; Murthy, S; Aertssen, S; Smits, JN; Nieczypor, P; Damen, E; Wegert, A; Nazaré, M; Lehtiö, L; Waaler, J; Krauss, S Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II. J Med Chem 64:17936-17949 (2021) [PubMed] Article More Info.:
Target
Name:
Protein mono-ADP-ribosyltransferase PARP3
Synonyms:
ADP-ribosyltransferase diphtheria toxin-like 3 | ADPRT-3 | ADPRT3 | ADPRTL3 | ARTD3 | IRT1 | NAD(+) ADP-ribosyltransferase 3 | PARP-3 | PARP3 | PARP3_HUMAN | Poly [ADP-ribose] polymerase 3 | Poly[ADP-ribose] synthase 3 | hPARP-3 | pADPRT-3
Type:
PROTEIN
Mol. Mass.:
60091.40
Organism:
Homo sapiens (Human)
Description:
ChEMBL_109545
Residue:
533
Sequence:
MAPKPKPWVQTEGPEKKKGRQAGREEDPFRSTAEALKAIPAEKRIIRVDPTCPLSSNPGTQVYEDYNCTLNQTNIENNNNKFYIIQLLQDSNRFFTCWNRWGRVGEVGQSKINHFTRLEDAKKDFEKKFREKTKNNWAERDHFVSHPGKYTLIEVQAEDEAQEAVVKVDRGPVRTVTKRVQPCSLDPATQKLITNIFSKEMFKNTMALMDLDVKKMPLGKLSKQQIARGFEALEALEEALKGPTDGGQSLEELSSHFYTVIPHNFGHSQPPPINSPELLQAKKDMLLVLADIELAQALQAVSEQEKTVEEVPHPLDRDYQLLKCQLQLLDSGAPEYKVIQTYLEQTGSNHRCPTLQHIWKVNQEGEEDRFQAHSKLGNRKLLWHGTNMAVVAAILTSGLRIMPHSGGRVGKGIYFASENSKSAGYVIGMKCGAHHVGYMFLGEVALGREHHINTDNPSLKSPPPGFDSVIARGHTEPDPTQDTELELDGQQVVVPQGQPVPCPEFSSSTFSQSEYLIYQESQCRLRYLLEVHL
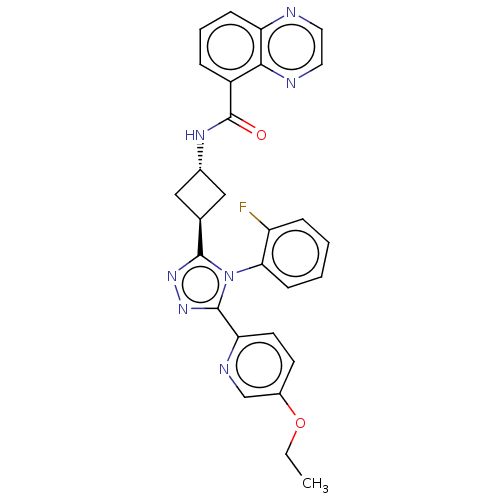
Inhibitor
Name:
BDBM50579974
Synonyms:
CHEMBL5090849 | US20230322721, Example 154 of WO 2019/243822
Type:
Small organic molecule
Emp. Form.:
C28H24FN7O2
Mol. Mass.:
509.5343
SMILES:
CCOc1ccc(nc1)-c1nnc([C@H]2C[C@@H](C2)NC(=O)c2cccc3nccnc23)n1-c1ccccc1F |r,wU:15.18,wD:13.13,(1.18,-57.78,;2.08,-56.54,;3.59,-56.7,;4.49,-55.47,;6.01,-55.63,;6.9,-54.39,;6.29,-53.01,;4.77,-52.84,;3.87,-54.07,;7.18,-51.78,;6.71,-50.31,;7.95,-49.39,;9.21,-50.31,;10.68,-49.82,;12.05,-50.53,;12.76,-49.16,;11.38,-48.45,;14.23,-48.68,;14.56,-47.17,;13.41,-46.13,;16.03,-46.68,;16.34,-45.17,;17.81,-44.69,;18.97,-45.72,;18.65,-47.24,;19.79,-48.27,;19.48,-49.77,;18,-50.26,;16.86,-49.22,;17.18,-47.72,;8.73,-51.78,;9.63,-53.02,;9,-54.41,;9.9,-55.64,;11.41,-55.48,;12.03,-54.08,;11.13,-52.85,;11.75,-51.45,)|