Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Mitogen-activated protein kinase 9
Ligand
BDBM50314070
Substrate
n/a
Meas. Tech.
ChEMBL_742476 (CHEMBL1769124)
Kd
16±n/a nM
Citation
 Goldstein, DM; Soth, M; Gabriel, T; Dewdney, N; Kuglstatter, A; Arzeno, H; Chen, J; Bingenheimer, W; Dalrymple, SA; Dunn, J; Farrell, R; Frauchiger, S; La Fargue, J; Ghate, M; Graves, B; Hill, RJ; Li, F; Litman, R; Loe, B; McIntosh, J; McWeeney, D; Papp, E; Park, J; Reese, HF; Roberts, RT; Rotstein, D; San Pablo, B; Sarma, K; Stahl, M; Sung, ML; Suttman, RT; Sjogren, EB; Tan, Y; Trejo, A; Welch, M; Weller, P; Wong, BR; Zecic, H Discovery of 6-(2,4-difluorophenoxy)-2-[3-hydroxy-1-(2-hydroxyethyl)propylamino]-8-methyl-8H-pyrido[2,3-d]pyrimidin-7-one (pamapimod) and 6-(2,4-difluorophenoxy)-8-methyl-2-(tetrahydro-2H-pyran-4-ylamino)pyrido[2,3-d]pyrimidin-7(8H)-one (R1487) as orally bioavailable and highly selective inhibitors J Med Chem 54:2255-65 (2011) [PubMed] Article
Goldstein, DM; Soth, M; Gabriel, T; Dewdney, N; Kuglstatter, A; Arzeno, H; Chen, J; Bingenheimer, W; Dalrymple, SA; Dunn, J; Farrell, R; Frauchiger, S; La Fargue, J; Ghate, M; Graves, B; Hill, RJ; Li, F; Litman, R; Loe, B; McIntosh, J; McWeeney, D; Papp, E; Park, J; Reese, HF; Roberts, RT; Rotstein, D; San Pablo, B; Sarma, K; Stahl, M; Sung, ML; Suttman, RT; Sjogren, EB; Tan, Y; Trejo, A; Welch, M; Weller, P; Wong, BR; Zecic, H Discovery of 6-(2,4-difluorophenoxy)-2-[3-hydroxy-1-(2-hydroxyethyl)propylamino]-8-methyl-8H-pyrido[2,3-d]pyrimidin-7-one (pamapimod) and 6-(2,4-difluorophenoxy)-8-methyl-2-(tetrahydro-2H-pyran-4-ylamino)pyrido[2,3-d]pyrimidin-7(8H)-one (R1487) as orally bioavailable and highly selective inhibitors J Med Chem 54:2255-65 (2011) [PubMed] Article More Info.:
Target
Name:
Mitogen-activated protein kinase 9
Synonyms:
JNK-55 | JNK2 | JNK2/JNK3 | MAPK9 | MK09_HUMAN | Mitogen-Activated Protein Kinase 9 (JNK2) | Mitogen-activated protein kinase 8/9 | PRKM9 | SAPK1A | Stress-activated protein kinase JNK2 | c-Jun N-terminal kinase 2 | c-Jun N-terminal kinase 2 (JNK2)
Type:
Enzyme
Mol. Mass.:
48131.49
Organism:
Homo sapiens (Human)
Description:
JNK-2 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology).
Residue:
424
Sequence:
MSDSKCDSQFYSVQVADSTFTVLKRYQQLKPIGSGAQGIVCAAFDTVLGINVAVKKLSRPFQNQTHAKRAYRELVLLKCVNHKNIISLLNVFTPQKTLEEFQDVYLVMELMDANLCQVIHMELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTACTNFMMTPYVVTRYYRAPEVILGMGYKENVDIWSVGCIMGELVKGCVIFQGTDHIDQWNKVIEQLGTPSAEFMKKLQPTVRNYVENRPKYPGIKFEELFPDWIFPSESERDKIKTSQARDLLSKMLVIDPDKRISVDEALRHPYITVWYDPAEAEAPPPQIYDAQLEEREHAIEEWKELIYKEVMDWEERSKNGVVKDQPSDAAVSSNATPSQSSSINDISSMSTEQTLASDTDSSLDASTGPLEGCR
Inhibitor
Name:
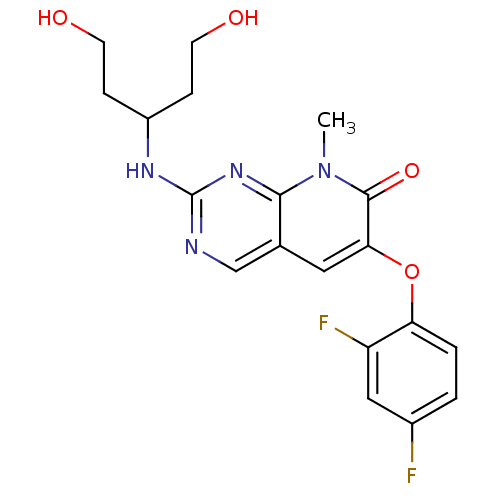
BDBM50314070
Synonyms:
6-(2,4-difluorophenoxy)-2-{[3-hydroxy-1-(2-hydroxyethyl)propyl]amino}-8-methylpyrido[2,3-d]pyrimidin-7(8H)-one | CHEMBL1090089 | pamapimod
Type:
Small organic molecule
Emp. Form.:
C19H20F2N4O4
Mol. Mass.:
406.3833
SMILES:
Cn1c2nc(NC(CCO)CCO)ncc2cc(Oc2ccc(F)cc2F)c1=O