Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Cholecystokinin receptor type A
Ligand
BDBM50005808
Substrate
n/a
Meas. Tech.
ChEBML_50060
IC50
1630±n/a nM
Citation
 Fincham, CI; Higginbottom, M; Hill, DR; Horwell, DC; O'Toole, JC; Ratcliffe, GS; Rees, DC; Roberts, E Amide bond replacements incorporated into CCK-B selective"dipeptoids". J Med Chem 35:1472-84 (1992) [PubMed] Article
Fincham, CI; Higginbottom, M; Hill, DR; Horwell, DC; O'Toole, JC; Ratcliffe, GS; Rees, DC; Roberts, E Amide bond replacements incorporated into CCK-B selective"dipeptoids". J Med Chem 35:1472-84 (1992) [PubMed] Article More Info.:
Target
Name:
Cholecystokinin receptor type A
Synonyms:
CCKAR_RAT | Cckar | Cholecystokinin peripheral | Cholecystokinin receptor | Cholecystokinin receptor type A
Type:
Enzyme Catalytic Domain
Mol. Mass.:
49676.37
Organism:
RAT
Description:
Cholecystokinin central 0 RAT::P30551
Residue:
444
Sequence:
MSHSPARQHLVESSRMDVVDSLLMNGSNITPPCELGLENETLFCLDQPQPSKEWQSALQILLYSIIFLLSVLGNTLVITVLIRNKRMRTVTNIFLLSLAVSDLMLCLFCMPFNLIPNLLKDFIFGSAVCKTTTYFMGTSVSVSTFNLVAISLERYGAICRPLQSRVWQTKSHALKVIAATWCLSFTIMTPYPIYSNLVPFTKNNNQTANMCRFLLPSDAMQQSWQTFLLLILFLLPGIVMVVAYGLISLELYQGIKFDASQKKSAKEKKPSTGSSTRYEDSDGCYLQKSRPPRKLELQQLSSGSGGSRLNRIRSSSSAANLIAKKRVIRMLIVIVVLFFLCWMPIFSANAWRAYDTVSAEKHLSGTPISFILLLSYTSSCVNPIIYCFMNKRFRLGFMATFPCCPNPGPPGVRGEVGEEEDGRTIRALLSRYSYSHMSTSAPPP
Inhibitor
Name:
BDBM50005808
Synonyms:
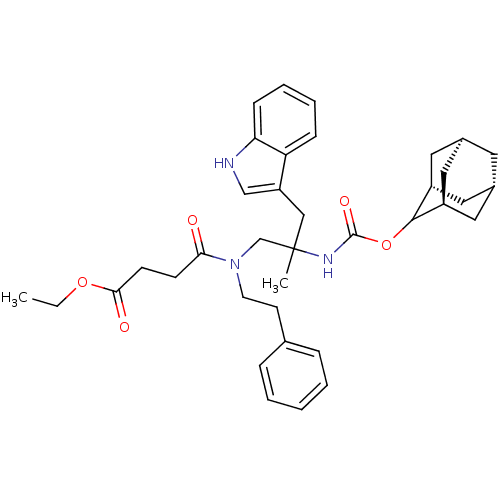
CHEMBL286664 | N-[2-(Adamantan-2-yloxycarbonylamino)-3-(1H-indol-3-yl)-2-methyl-propyl]-N-phenethyl-succinamic acid ethyl ester
Type:
Small organic molecule
Emp. Form.:
C37H47N3O5
Mol. Mass.:
613.7862
SMILES:
CCOC(=O)CCC(=O)N(CCc1ccccc1)CC(C)(Cc1c[nH]c2ccccc12)NC(=O)OC1[C@H]2C[C@@H]3C[C@@H](C[C@H]1C3)C2 |wU:40.48,wD:38.40,42.44,36.49,TLB:34:35:41:38.43.39,THB:37:36:41:38.43.39,37:38:41:35.36.44,39:40:35:38.43.37,(17.79,-15.28,;17.38,-13.79,;15.9,-13.4,;15.51,-11.91,;16.6,-10.82,;14.01,-11.51,;13.62,-10.02,;12.14,-9.61,;11.05,-10.7,;11.73,-8.13,;12.82,-7.04,;14.3,-7.43,;15.39,-6.34,;14.97,-4.86,;16.05,-3.77,;17.54,-4.15,;17.96,-5.63,;16.87,-6.72,;10.86,-6.85,;9.35,-6.98,;10.11,-8.29,;8.1,-6.08,;8.26,-4.54,;7.11,-3.51,;7.72,-2.1,;9.26,-2.26,;10.38,-1.22,;11.86,-1.68,;12.18,-3.19,;11.05,-4.24,;9.58,-3.77,;8.57,-8.29,;7.03,-8.28,;6.27,-6.94,;6.24,-9.6,;4.7,-9.57,;4.7,-11.09,;3.67,-12.37,;2.28,-11.79,;.78,-12.21,;1.97,-10.93,;1.97,-9.45,;3.32,-8.99,;2.26,-10.22,;3.29,-11.44,)|