Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Bifunctional purine biosynthesis protein ATIC
Ligand
BDBM24686
Substrate
n/a
Meas. Tech.
Enzyme Activity Assays
Ki
>1.00e+5±n/a nM
Citation
 Connelly, S; DeMartino, JK; Boger, DL; Wilson, IA Biological and structural evaluation of 10R- and 10S-methylthio-DDACTHF reveals a new role for sulfur in inhibition of glycinamide ribonucleotide transformylase. Biochemistry 52:5133-44 (2013) [PubMed] Article
Connelly, S; DeMartino, JK; Boger, DL; Wilson, IA Biological and structural evaluation of 10R- and 10S-methylthio-DDACTHF reveals a new role for sulfur in inhibition of glycinamide ribonucleotide transformylase. Biochemistry 52:5133-44 (2013) [PubMed] Article More Info.:
Target
Name:
Bifunctional purine biosynthesis protein ATIC
Synonyms:
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase | 5-aminoimidazole-4-carboxamide-ribonucleotide transformylase | AICAR Tfase | AICAR transformylase | ATIC | Aminoimidazole carboxamide ribonucleotide transformylase (AICAR Tfase) | Bifunctional purine biosynthesis protein PURH | IMP Cyclohydrolase (IMPCH) | IMP cyclohydrolase | IMP synthetase | Inosinicase | PUR9_HUMAN | PURH | Phosphoribosylaminoimidazolecarboxamide formyltransferase | Thymidylate synthase/GAR transformylase/AICAR transformylase
Type:
Protein
Mol. Mass.:
64616.62
Organism:
Homo sapiens (Human)
Description:
P31939
Residue:
592
Sequence:
MAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARLDFNLIRVVACNLYPFVKTVASPGVTVEEAVEQIDIGGVTLLRAAAKNHARVTVVCEPEDYVVVSTEMQSSESKDTSLETRRQLALKAFTHTAQYDEAISDYFRKQYSKGVSQMPLRYGMNPHQTPAQLYTLQPKLPITVLNGAPGFINLCDALNAWQLVKELKEALGIPAAASFKHVSPAGAAVGIPLSEDEAKVCMVYDLYKTLTPISAAYARARGADRMSSFGDFVALSDVCDVPTAKIISREVSDGIIAPGYEEEALTILSKKKNGNYCVLQMDQSYKPDENEVRTLFGLHLSQKRNNGVVDKSLFSNVVTKNKDLPESALRDLIVATIAVKYTQSNSVCYAKNGQVIGIGAGQQSRIHCTRLAGDKANYWWLRHHPQVLSMKFKTGVKRAEISNAIDQYVTGTIGEDEDLIKWKALFEEVPELLTEAEKKEWVEKLTEVSISSDAFFPFRDNVDRAKRSGVAYIAAPSGSAADKVVIEACDELGIILAHTNLRLFHH
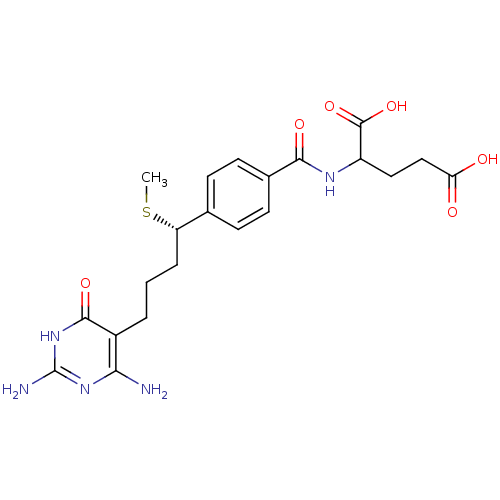
Inhibitor
Name:
BDBM24686
Synonyms:
10-thiomethyl-DDACTHF, 10S-3 | 10S (7) | 2-({4-[(1S)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}formamido)pentanedioic acid
Type:
Small organic molecule
Emp. Form.:
C21H27N5O6S
Mol. Mass.:
477.534
SMILES:
CS[C@@H](CCCc1c(N)nc(N)[nH]c1=O)c1ccc(cc1)C(=O)NC(CCC(O)=O)C(O)=O |r|