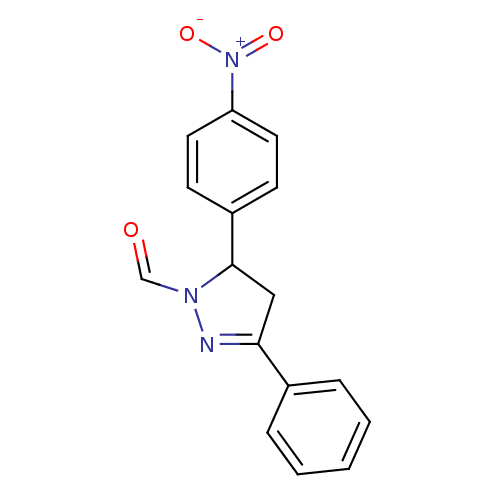
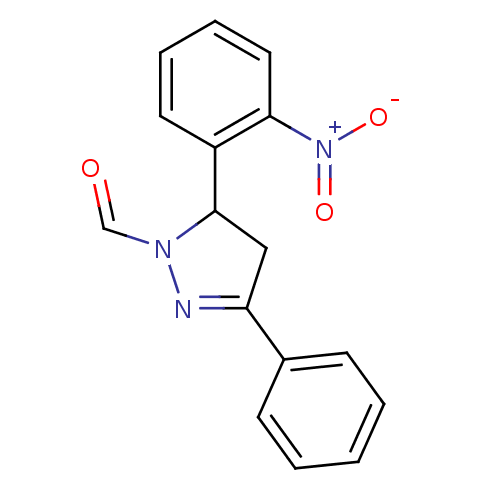
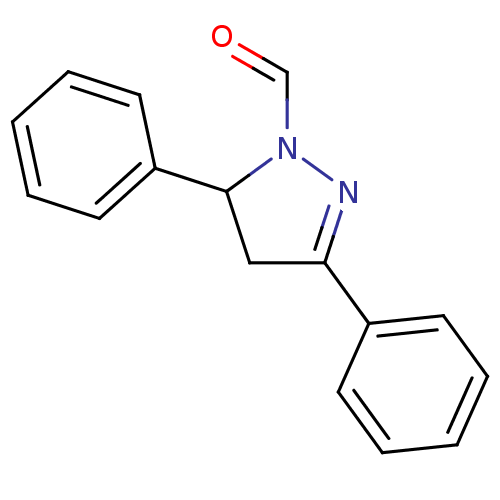
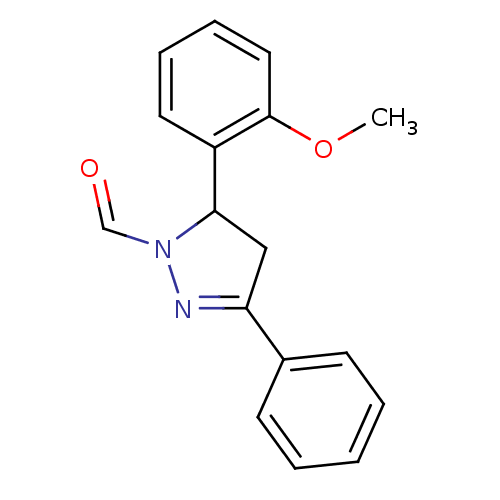
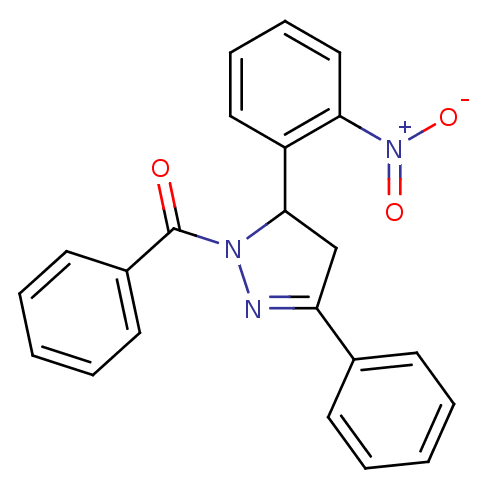
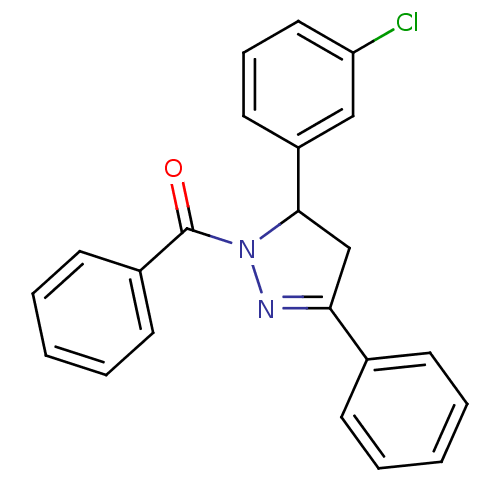
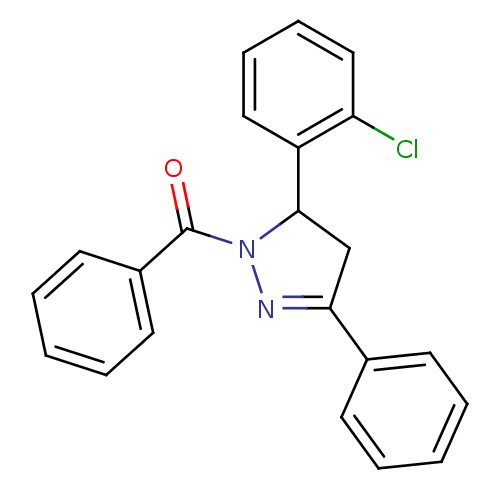
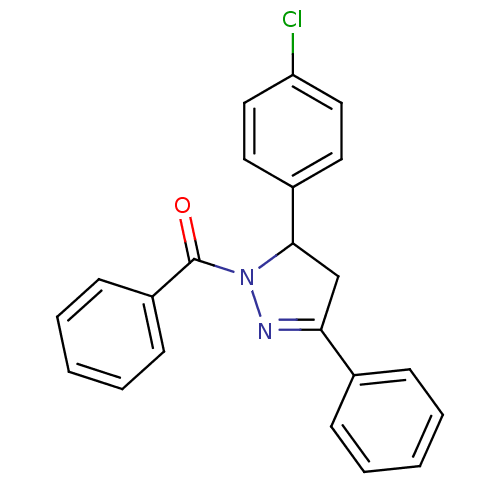
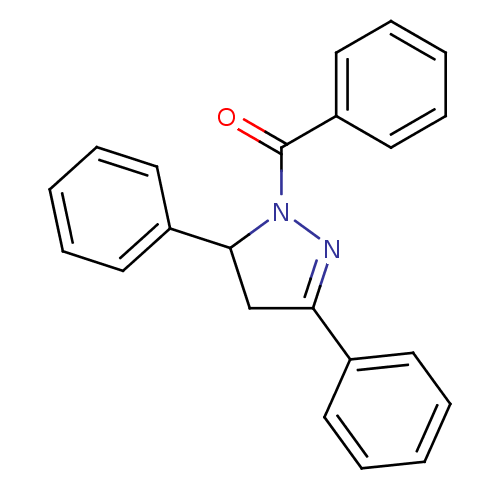
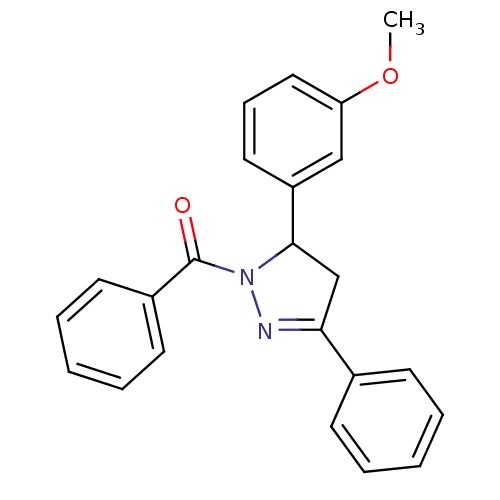
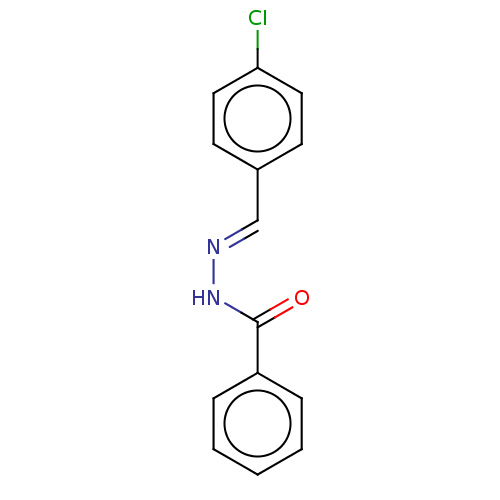
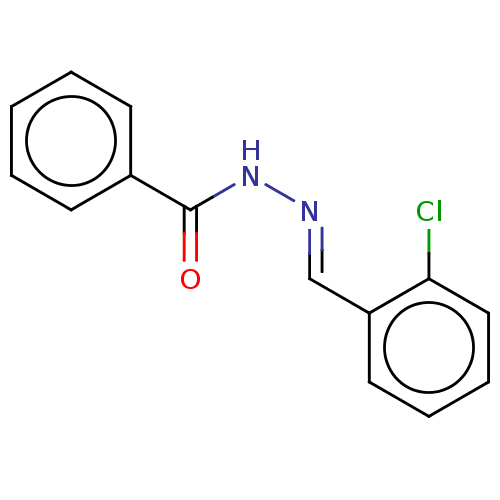
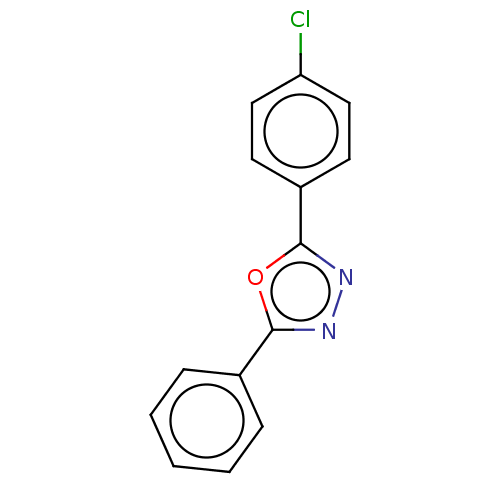
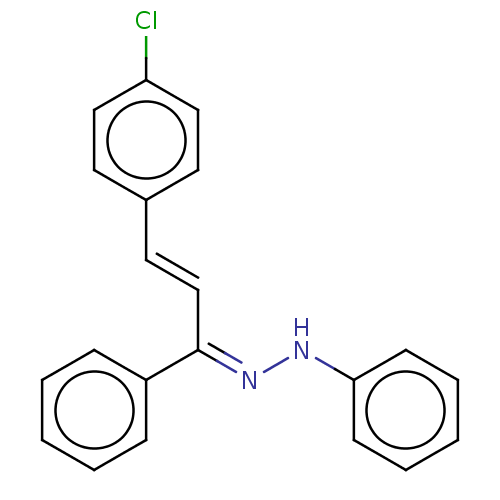
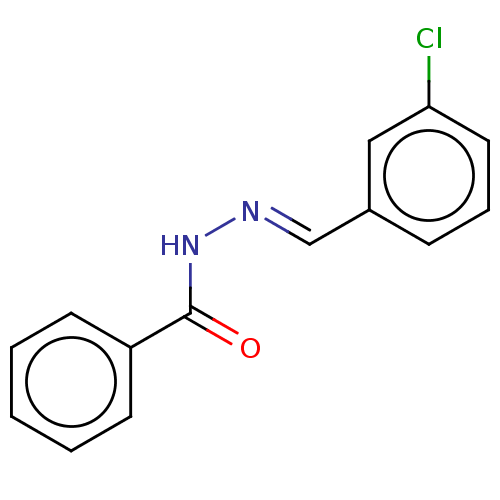
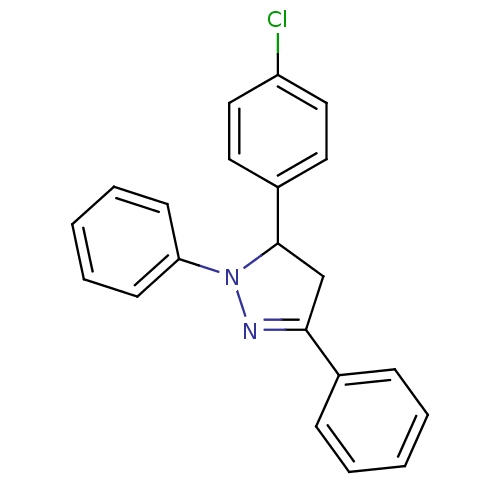
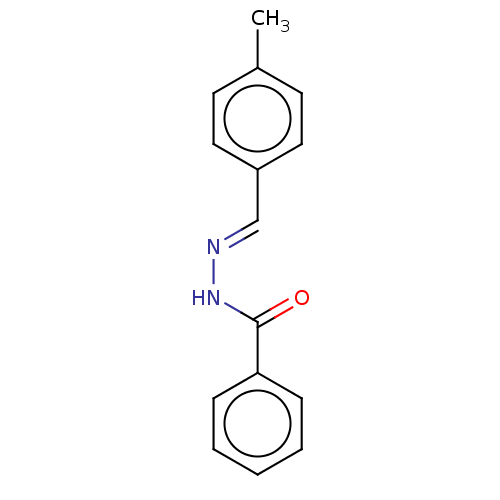
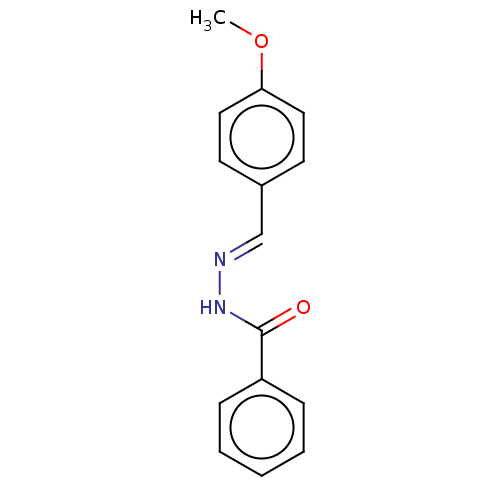
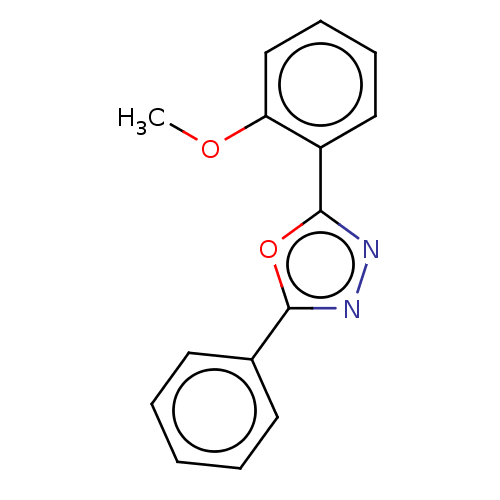
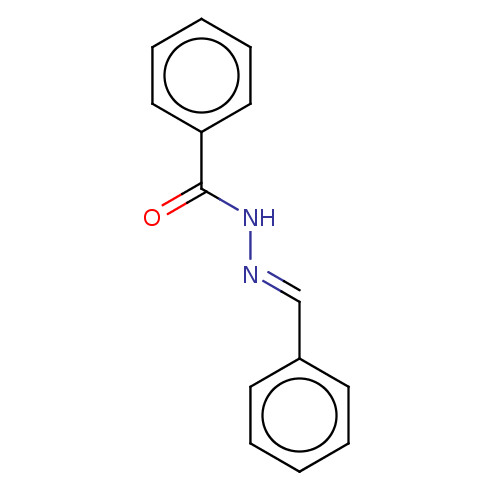
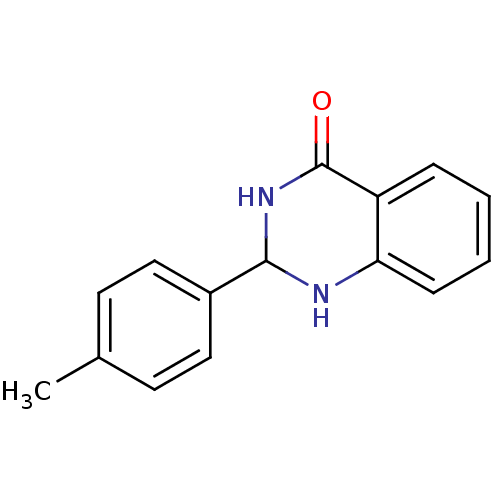
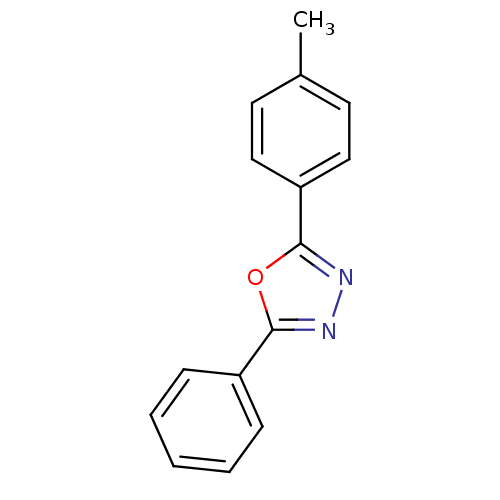
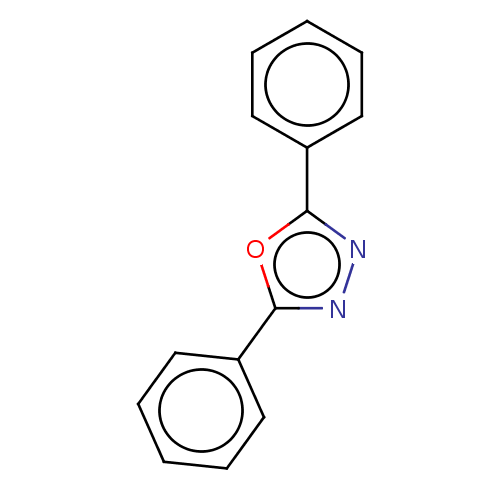
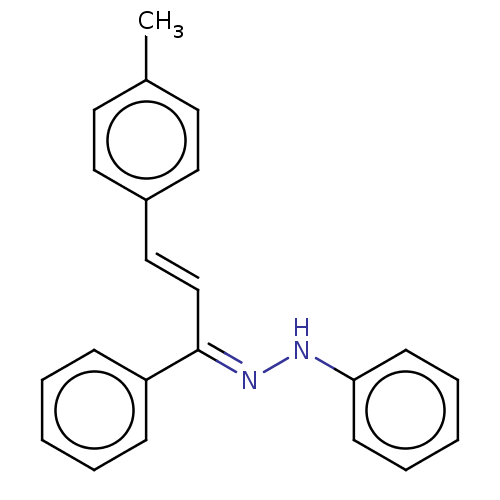
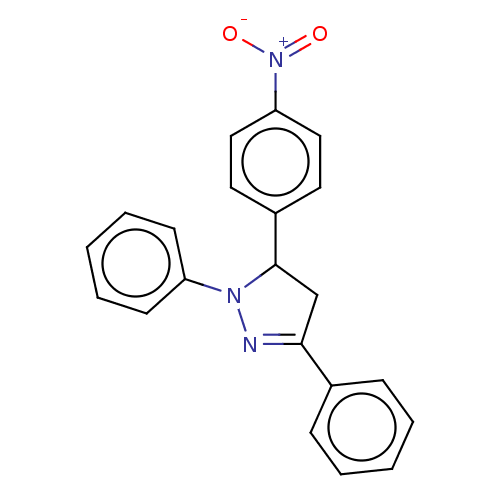
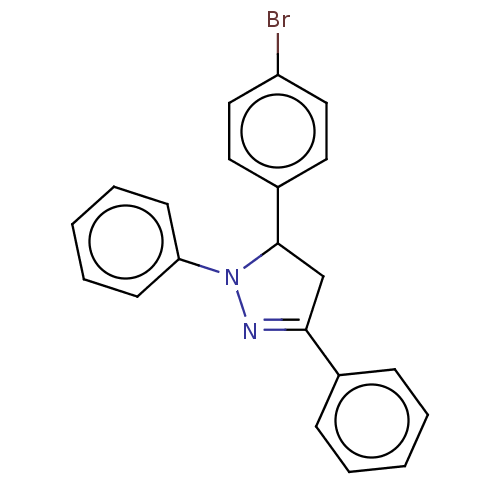
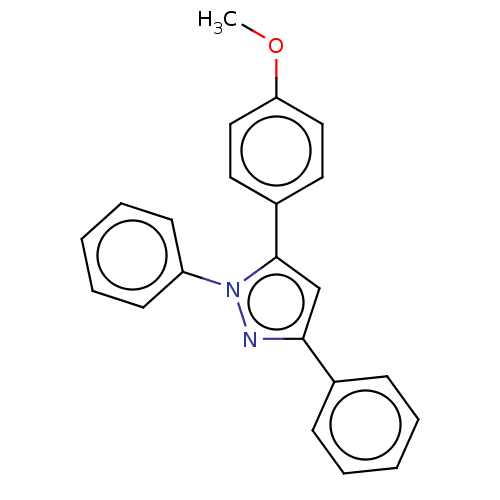
Affinity DataKi: 0.519nM ΔG°: -13.2kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 0.580nM ΔG°: -13.1kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 0.963nM ΔG°: -12.8kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 1.30nM ΔG°: -12.6kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 1.35nM ΔG°: -12.6kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 1.37nM ΔG°: -12.6kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 1.74nM ΔG°: -12.4kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 2.21nM ΔG°: -12.3kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 2.72nM ΔG°: -12.1kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 3.87nM ΔG°: -11.9kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 9.80nM ΔG°: -11.4kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 12.7nM ΔG°: -11.2kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 14.4nM ΔG°: -11.1kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 18.1nM ΔG°: -11.0kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 18.9nM ΔG°: -11.0kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 30.1nM ΔG°: -10.7kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 32nM ΔG°: -10.6kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 41nM ΔG°: -10.5kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 50.3nM ΔG°: -10.4kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 96.5nM ΔG°: -9.95kcal/molepH: 6.0 T: 2°CAssay Description:Enzyme was equilibrated in 0.1 M phosphate buffer, pH 6.0 at 37 °C. All compounds were prepared in DMSO. Various concentrations of compounds we...More data for this Ligand-Target Pair
Affinity DataKi: 440nM ΔG°: -9.01kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 560nM ΔG°: -8.87kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 680nM ΔG°: -8.75kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.00E+3nM ΔG°: -8.51kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.11E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.13E+3nM ΔG°: -8.43kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.13E+3nM ΔG°: -8.43kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.17E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.26E+3nM ΔG°: -8.37kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.33E+3nM ΔG°: -8.33kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.33E+3nM ΔG°: -8.33kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+3nM ΔG°: -8.30kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.47E+3nM ΔG°: -8.27kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.56E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.61E+3nM ΔG°: -8.22kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.68E+3nM ΔG°: -8.19kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 1.70E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.91E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.12E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.14E+3nM ΔG°: -8.04kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 2.35E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.50E+3nMpH: 6.0Assay Description:Cathepsin B and H were first activated in the presence of thiol activators at pH 6.0 and pH 7.0, respectively. Then, 15 µL of the enzyme solution was...More data for this Ligand-Target Pair
Affinity DataKi: 2.55E+3nM ΔG°: -7.93kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 2.58E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.66E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.72E+3nM ΔG°: -7.89kcal/molepH: 5.0 T: 2°CAssay Description:The proteolytic activity was estimated at pH 5.0, 37 °C using 0.1 M acetate buffer as the incubation medium. The homogenate prepared above was incuba...More data for this Ligand-Target Pair
Affinity DataKi: 2.91E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 2.91E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 3.16E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 3.33E+3nMAssay Description:Competitive inhibition of goat liver cathepsin H using Leu-betaNA substrate by Lineweaver-Burk plot analysisMore data for this Ligand-Target Pair