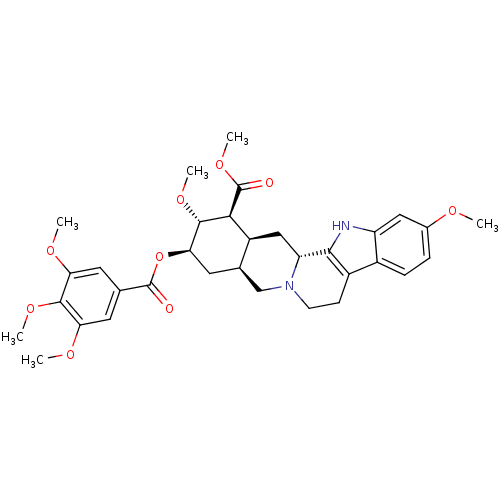
BDBM50017712 (-)-reserpine::(3beta,16beta,17alpha,18beta,20alpha)-11,17-dimethoxy-18-[(3,4,5-trimethoxybenzoyl)oxy]yohimban-16-carboxylic acid methyl ester::3,4,5-trimethoxybenzoyl methyl reserpate::Apoplon::CHEMBL772::NCGC00091250::RESERPINE::Reserpin::Serpalan::cid_5770::methyl (3beta,16beta,17alpha,18beta,20alpha)-11,17-dimethoxy-18-[(3,4,5-trimethoxybenzoyl)oxy]yohimban-16-carboxylate
SMILES CO[C@H]1[C@@H](C[C@@H]2CN3CCc4c([nH]c5cc(OC)ccc45)[C@H]3C[C@@H]2[C@@H]1C(=O)OC)OC(=O)c1cc(OC)c(OC)c(OC)c1
InChI Key InChIKey=QEVHRUUCFGRFIF-MDEJGZGSSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 68 hits for monomerid = 50017712
Found 68 hits for monomerid = 50017712
Affinity DataKi: 1nMAssay Description:Inhibition of dopamine uptake at VMAT in bovine chromaffin granule ghostsMore data for this Ligand-Target Pair
Affinity DataKi: 5.30nMAssay Description:Displacement of [3H]reserpine from human VMAT2 expressed in HEK293 cell membranes incubated for 60 mins by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 100nMAssay Description:Concentration giving half of the maximal ATPase activity calculated for the high-affinity binding site of the CHO P-Glycoprotein (P-gp) in two-affini...More data for this Ligand-Target Pair
Affinity DataKi: 410nMAssay Description:Displacement of [3H](+)-syn-Ethyl 1-(2-(2,4-Dioxo-1,2-dihydroquinazolin-3(4H)-yl)ethyl)-4-(4-fluorophenyl)piperidine-3-carboxylate from human VMAT2 e...More data for this Ligand-Target Pair
Affinity DataKi: 630nMAssay Description:Displacement of [3H]DHTB from human VMAT2 expressed in HEK293 cell membranes incubated for 90 mins by microbeta scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 970nMAssay Description:TP_TRANSPORTER: increase in Vinblastine intracellular accumulation in MDR1-expressing LLC-PK1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+3nMAssay Description:Displacement of [125I]DOI from human 5HT2A receptor expressed in HEK293 cell membranesMore data for this Ligand-Target Pair
Affinity DataKi: 2.31E+3nMAssay Description:TP_TRANSPORTER: transepithelial transport of digoxin (basal to apical) in Caco-2 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 4.03E+3nMAssay Description:TP_TRANSPORTER: increase in Calcein-AM intracellular accumulation in mdr1a-expressing LLC-PK1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.15E+4nMAssay Description:TP_TRANSPORTER: inhibition of Digoxin transepithelial transport (basal to apical) in MDR1-expressing MDCK cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.22E+4nMAssay Description:TP_TRANSPORTER: increase in Calcein-AM intracellular accumulation in MDR1-expressing LLC-PK1 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.79E+4nMAssay Description:TP_TRANSPORTER: increase in Calcein-AM intracellular accumulation in mdr1b-expressing LLC-PK1 cellsMore data for this Ligand-Target Pair
TargetSolute carrier family 22 member 1(Rat)
Bayerische Julius-Maximilians-Universit£T
Curated by ChEMBL
Bayerische Julius-Maximilians-Universit£T
Curated by ChEMBL
Affinity DataKi: >2.00E+4nMAssay Description:TP_TRANSPORTER: inhibition of TEA uptake in Xenopus laevis oocytesMore data for this Ligand-Target Pair
Affinity DataKi: 2.95E+5nMAssay Description:TP_TRANSPORTER: inhibition of Vinblastine transepithelial transport (basal to apical) in MRP2-expressing MDCK cellsMore data for this Ligand-Target Pair
TargetQuinolone resistance protein NorA(Staphylococcus aureus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataIC50: 8.20E+3nMAssay Description:Inhibition of norA-mediated ethidium bromide efflux in methicillin-resistant Staphylococcus aureus SA-1199B by spectrofluorometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.38E+4nMT: 25°CAssay Description:Briefly, 3 uL of enzyme (approximately 20 nM LoxA, final concentration) or buffer (no-enzyme control) was dispensed into 1536-well Greiner black clea...More data for this Ligand-Target Pair
TargetCystic fibrosis transmembrane conductance regulator(Human)
Southern Research Institute
Curated by PubChem BioAssay
Southern Research Institute
Curated by PubChem BioAssay
Affinity DataIC50: 2.5nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 4.5nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 2nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 4.90nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 4.30nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 1.30nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: 3.70nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: >50nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
Affinity DataIC50: >100nMpH: 8.5Assay Description:The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM Tricine, 10 mM Tris, and 5 mM MgCl2, pH 8.5. Liposomes (1 _...More data for this Ligand-Target Pair
TargetQuinolone resistance protein NorA(Staphylococcus aureus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataIC50: 7.00E+3nMAssay Description:Inhibition of NorA in Staphylococcus aureus 1199B assessed as inhibition of ethidium bromide efflux dose response curve based fluorometric assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+3nMAssay Description:Inhibition of binding of Batrachotoxinin [3H]BTX-B to high affinity sites on voltage dependent sodium channels in a vesicular preparation from guinea...More data for this Ligand-Target Pair
Affinity DataIC50: 3.02E+3nMAssay Description:Inhibition of P-glycoprotein by Hoechst assayMore data for this Ligand-Target Pair
Affinity DataIC50: 3.24E+3nMAssay Description:Inhibition of P-glycoprotein expressed in A2780/ADR cells by calcein AM assayMore data for this Ligand-Target Pair
Affinity DataIC50: 3.90E+3nMAssay Description:TP_TRANSPORTER: inhibition of Rhodamine 123 efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 5.30E+3nMAssay Description:TP_TRANSPORTER: inhibition of Tetramethylrosamine efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
TargetQuinolone resistance protein NorA(Staphylococcus aureus)
University of Copenhagen
Curated by ChEMBL
University of Copenhagen
Curated by ChEMBL
Affinity DataIC50: 9.20E+3nMAssay Description:Inhibition of NorA in Staphylococcus aureus 1199B harboring grlA A116E mutant assessed as inhibition of ethidium bromide efflux measured for 5 mins b...More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:TP_TRANSPORTER: inhibition of Daunorubicin efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:TP_TRANSPORTER: inhibition of Fluo-3-AM efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:TP_TRANSPORTER: inhibition of LDS-751 efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 6.10E+3nMAssay Description:TP_TRANSPORTER: inhibition of JC-1 efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 2.60E+3nMAssay Description:TP_TRANSPORTER: inhibition of Calcein-AM efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 1.02E+4nMAssay Description:TP_TRANSPORTER: increase in dihydrofluorescein intracellular accumulation (dihydrofluorescein: 1 uM) in SK-E2 cells (expressing BSEP)More data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+5nMAssay Description:Inhibition of human recombinant DNA topoisomerase1More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)