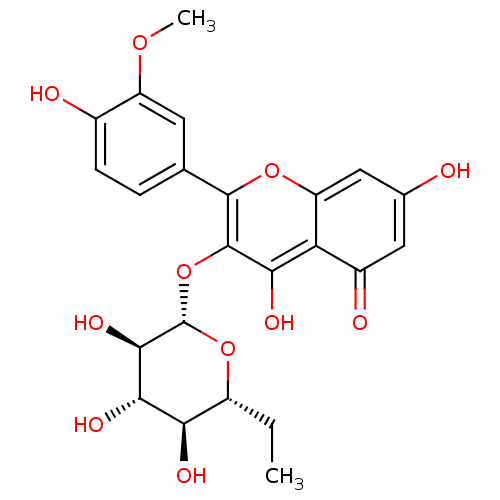
BDBM108033 3'-methoxyhirsutrin (M7)::3‐{[(2S,3R,4S,5S,6R)‐6‐ethyl‐3,4,5‐trihydroxyoxan‐2‐ yl]oxy}‐5,7‐dihydroxy‐2‐(4‐hydroxy‐3‐ methoxyphenyl)‐4H‐chromen‐4‐one
SMILES CC[C@H]1O[C@@H](Oc2c(O)c3c(cc(O)cc3=O)oc2-c2ccc(O)c(OC)c2)[C@H](O)[C@@H](O)[C@@H]1O
InChI Key InChIKey=PXFNLQXLPBSCDQ-KDLFCHMISA-N
Data 2 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 108033
Found 2 hits for monomerid = 108033
Affinity DataIC50: 2.46E+5nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 8.60E+3nMpH: 7.6 T: 2°CAssay Description:Inhibition of xanthine oxidase (XO) by each isolated phenolics was measured by following the decrease in the uric acid formation at 293nm at 25°...More data for this Ligand-Target Pair