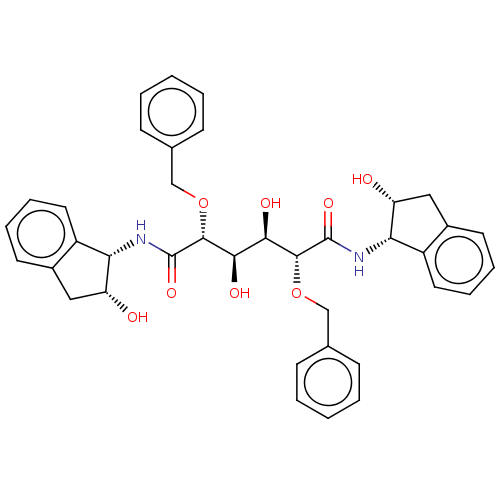
BDBM358 (2R,3R,4R,5R)-2,5-bis(benzyloxy)-3,4-dihydroxy-N,N'-bis[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]hexanediamide::1-Valine Methylamide deriv. 11::C2-symmetric compound 8::Diol-Based HIV-1 protease inhibitor 1::symmetric/asymmetric inhibitor 2
SMILES O[C@H]([C@@H](O)[C@@H](OCc1ccccc1)C(=O)N[C@@H]1[C@H](O)Cc2ccccc12)[C@@H](OCc1ccccc1)C(=O)N[C@@H]1[C@H](O)Cc2ccccc12
InChI Key InChIKey=UOHMQWQGAJAUGT-JQFCFGFHSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 6 hits for monomerid = 358
Found 6 hits for monomerid = 358
Affinity DataKi: 0.200nM ΔG°: -13.4kcal/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 0.600nM ΔG°: -12.8kcal/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 1.22nM ΔG°: -12.4kcal/molepH: 5.0 T: 2°CAssay Description:A fluorimetric assay was used to determine the effects of the inhibitors on HIV-1 protease. This assay used an internally quenched fluorescent peptid...More data for this Ligand-Target Pair
Affinity DataKi: 25nMAssay Description:Enzyme activities were assayed by monitoring the hydrolysis of substrate in the presence or absence of inhibitor compounds. The hydrolysis was record...More data for this Ligand-Target Pair
Affinity DataKi: 85nM ΔG°: -9.54kcal/molepH: 4.5 T: 2°CAssay Description:Enzyme activities were assayed by monitoring the hydrolysis of substrate in the presence or absence of inhibitor compounds. The hydrolysis was record...More data for this Ligand-Target Pair
Affinity DataKi: >2.00E+3nMAssay Description:Enzyme activities were assayed by monitoring the hydrolysis of substrate in the presence or absence of inhibitor compounds. The hydrolysis was record...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)