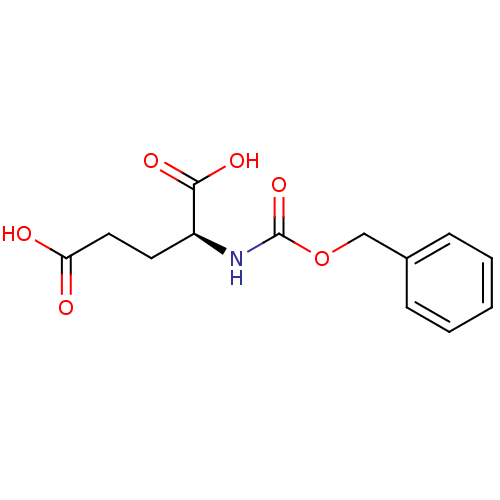
BDBM36120 Cbz-L-Glu::Z-L-Glu
SMILES OC(=O)CC[C@H](NC(=O)OCc1ccccc1)C(O)=O
InChI Key InChIKey=PVFCXMDXBIEMQG-JTQLQIEISA-N
Data 7 ITC
Activity Spreadsheet -- ITC Data from BindingDB
 Found 7 hits for monomerid = 36120
Found 7 hits for monomerid = 36120
ITC DataΔG°: -1.53kcal/mole −TΔS°: 3.58kcal/mole ΔH°: -5.02kcal/mole logk: 14
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
ITC Data−TΔS°: -0.128kcal/mole ΔH°: -2.51kcal/mole logk: 86
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.498kcal/mole ΔH°: -2.51kcal/mole logk: 162
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.242kcal/mole ΔH°: -2.60kcal/mole logk: 119
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.904kcal/mole ΔH°: -1.86kcal/mole logk: 107
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.762kcal/mole ΔH°: -2.13kcal/mole logk: 133
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.0498kcal/mole ΔH°: -3.05kcal/mole logk: 189
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails