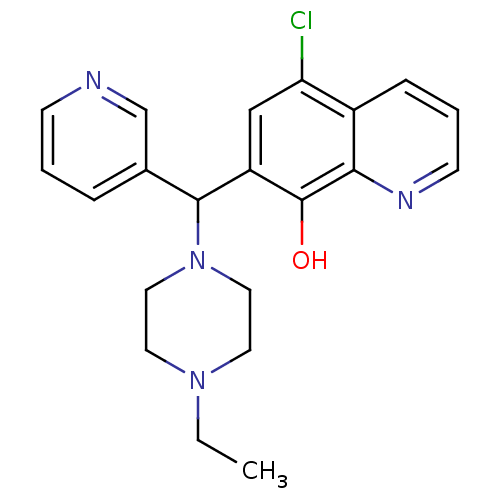
BDBM49152 5-chloranyl-7-[(4-ethylpiperazin-1-yl)-pyridin-3-yl-methyl]quinolin-8-ol::5-chloro-7-[(4-ethyl-1-piperazinyl)-(3-pyridinyl)methyl]-8-quinolinol::5-chloro-7-[(4-ethylpiperazin-1-yl)-pyridin-3-ylmethyl]quinolin-8-ol::5-chloro-7-[(4-ethylpiperazino)-(3-pyridyl)methyl]quinolin-8-ol::BRD4354::MLS000564806::SMR000151966::cid_3516032
SMILES CCN1CCN(CC1)C(c1cccnc1)c1cc(Cl)c2cccnc2c1O
InChI Key InChIKey=XAFSFXYJOJWLJD-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 12 hits for monomerid = 49152
Found 12 hits for monomerid = 49152
Affinity DataIC50: 2.39E+3nMAssay Description:NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: John A. Katzenellenbogen, ...More data for this Ligand-Target Pair
TargetNeutrophil cytosol factor 1 [S99G](Homo sapiens (Human))
Emory University
Curated by PubChem BioAssay
Emory University
Curated by PubChem BioAssay
Affinity DataIC50: 5.00E+4nMAssay Description:NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Susan Smith, Emory Univers...More data for this Ligand-Target Pair
Affinity DataEC50: >1.95E+5nMAssay Description:Keywords: Heat Shock Factor-1 (HSF-1), Stress Response, MG132, NIH3T3, Luminescence Assay Overview: Modified NIH3T3, transformed to express firefly...More data for this Ligand-Target Pair
TargetMitochondrial import inner membrane translocase subunit TIM10(Saccharomyces cerevisiae S288c)
Burnham Center For Chemical Genomics
Curated by PubChem BioAssay
Burnham Center For Chemical Genomics
Curated by PubChem BioAssay
Affinity DataIC50: 1.80E+4nMAssay Description:Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA...More data for this Ligand-Target Pair
Affinity DataIC50: 4.04E+4nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 1.88E+3nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 6.51E+4nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 1.38E+4nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 850nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 8.44E+3nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 3.89E+3nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair
Affinity DataIC50: 6.72E+4nMpH: 7.4Assay Description:Briefly, this fluorogenic assays uses an acetylated lysine tripeptide substrate, amide-linked to a fluorescently quenched aminocoumarin (AMC). Enzyme...More data for this Ligand-Target Pair