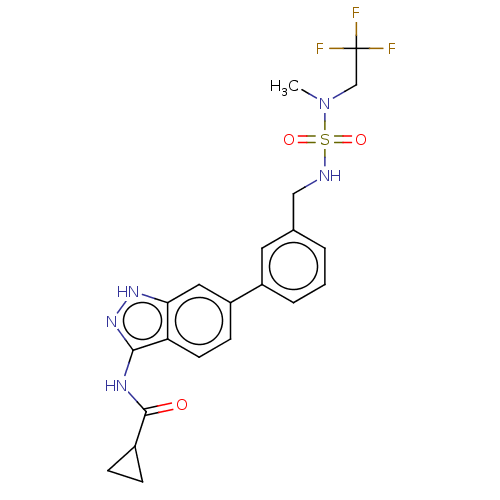
BDBM50511384 CHEMBL4561238
SMILES CN(CC(F)(F)F)S(=O)(=O)NCc1cccc(c1)-c1ccc2c(NC(=O)C3CC3)n[nH]c2c1
InChI Key InChIKey=MCYCSSJHDPRHBZ-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 8 hits for monomerid = 50511384
Found 8 hits for monomerid = 50511384
TargetBMP-2-inducible protein kinase(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataKi: 42nMAssay Description:Displacement of Alexafluor labelled kinase tracer236 from biotinylated C-terminal Avi-tagged BMP2K kinase domain (38 to 345 residues) (unknown origin...More data for this Ligand-Target Pair
TargetAP2-associated protein kinase 1(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataKi: 44nMAssay Description:Displacement of Alexafluor labelled kinase tracer236 from biotinylated C-terminal Avi-tagged AAK1 kinase domain (31 to 396 residues) (unknown origin)...More data for this Ligand-Target Pair
TargetSerine/threonine-protein kinase 16(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataKi: 290nMAssay Description:Displacement of Alexafluor labelled kinase tracer236 from biotinylated C-terminal Avi-tagged STK16 kinase domain (13 to 305 residues) (unknown origin...More data for this Ligand-Target Pair
TargetCyclin-G-associated kinase(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataKi: 780nMAssay Description:Displacement of Alexafluor labelled kinase tracer236 from biotinylated C-terminal Avi-tagged GAK kinase domain (12 to 347 residues) (unknown origin) ...More data for this Ligand-Target Pair
TargetSerine/threonine-protein kinase 16(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataIC50: >1.00E+4nMAssay Description:Inhibition of tracer K5 binding to N-terminal nano luciferase-fused STK16 (unknown origin) expressed in HEK293 cells incubated for 2 hrs followed by ...More data for this Ligand-Target Pair
TargetAP2-associated protein kinase 1(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataIC50: 3.40E+3nMAssay Description:Inhibition of tracer K5 binding to N-terminal nano luciferase-fused AAK1 (unknown origin) expressed in HEK293 cells incubated for 2 hrs followed by N...More data for this Ligand-Target Pair
TargetBMP-2-inducible protein kinase(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataIC50: 3.03E+3nMAssay Description:Inhibition of tracer K5 binding to N-terminal nano luciferase-fused BMP2K (unknown origin) expressed in HEK293 cells incubated for 2 hrs followed by ...More data for this Ligand-Target Pair
TargetCyclin-G-associated kinase(Homo sapiens (Human))
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
University Of North Carolina At Chapel Hill (Unc-Ch)
Curated by ChEMBL
Affinity DataIC50: >1.00E+4nMAssay Description:Inhibition of tracer K5 binding to N-terminal nano luciferase-fused GAK (unknown origin) expressed in HEK293 cells incubated for 2 hrs followed by Na...More data for this Ligand-Target Pair