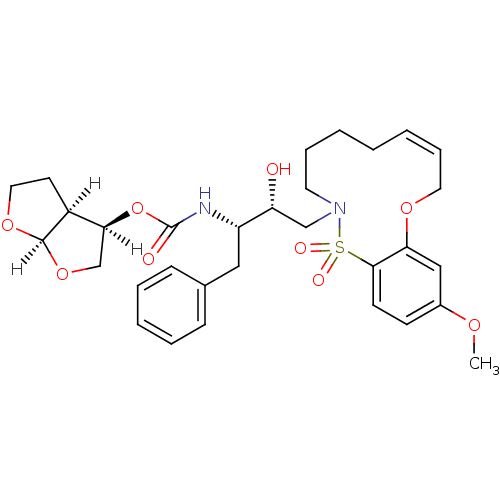
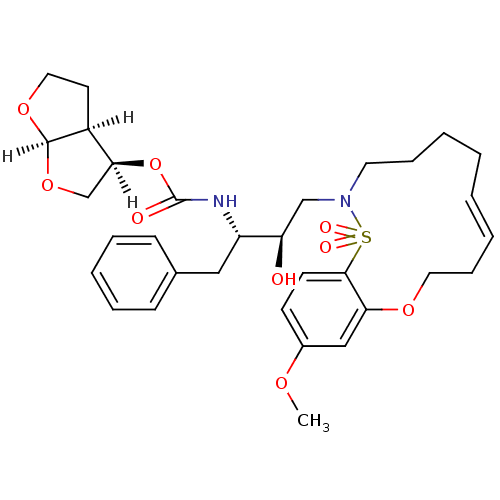
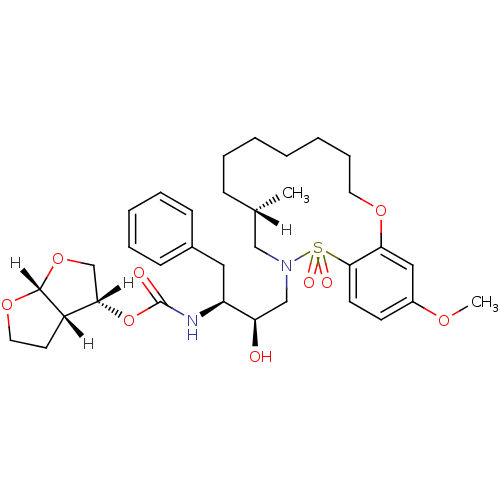
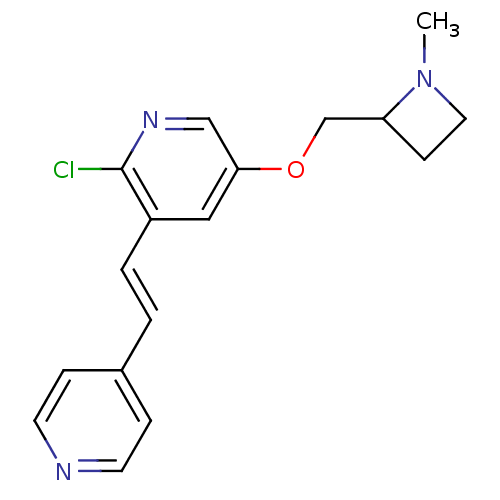
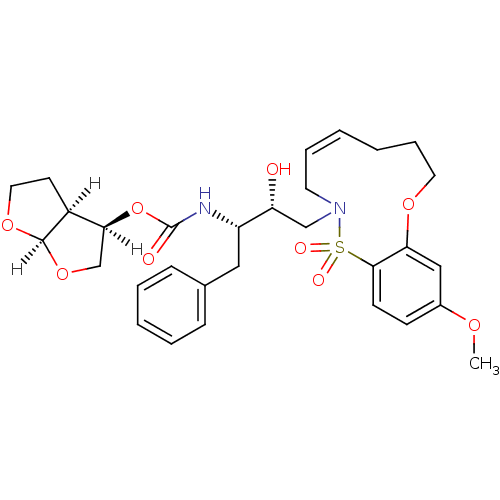
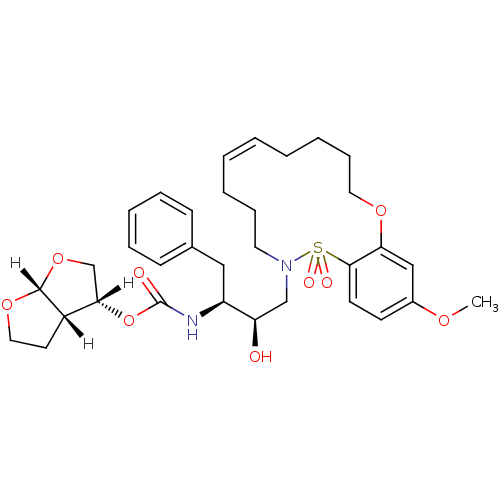
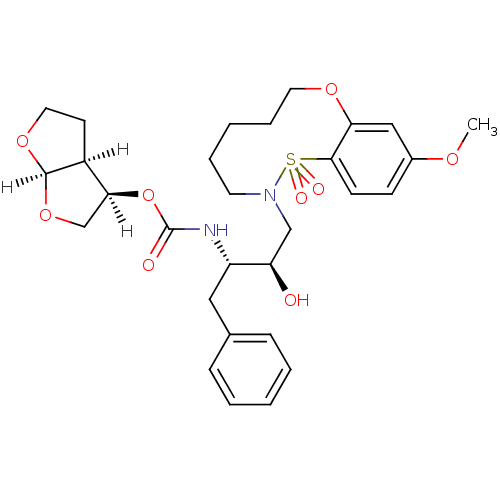
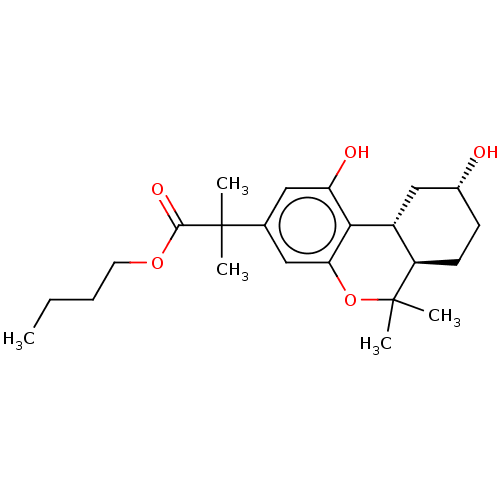
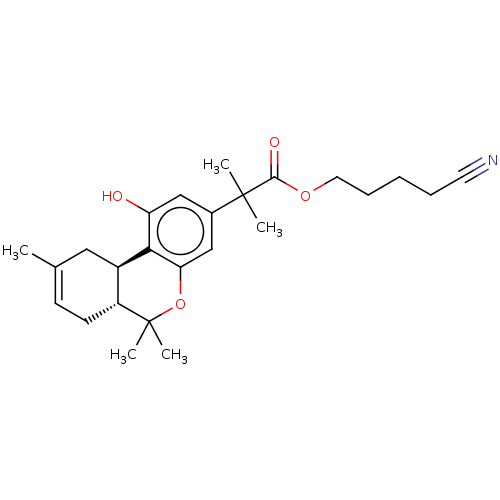
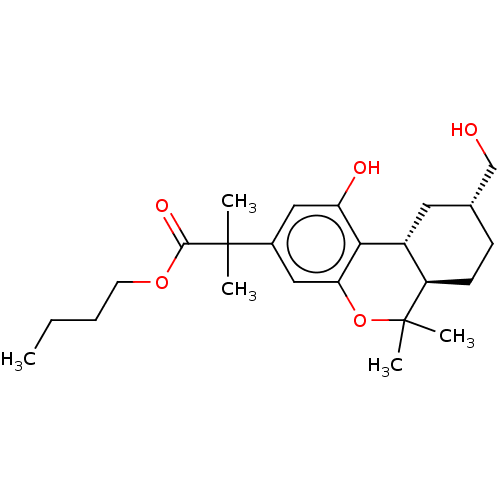
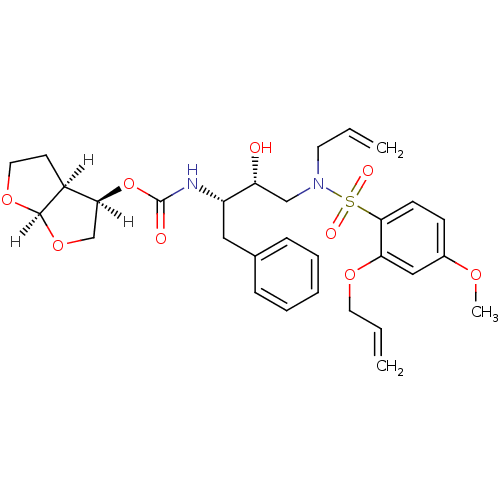
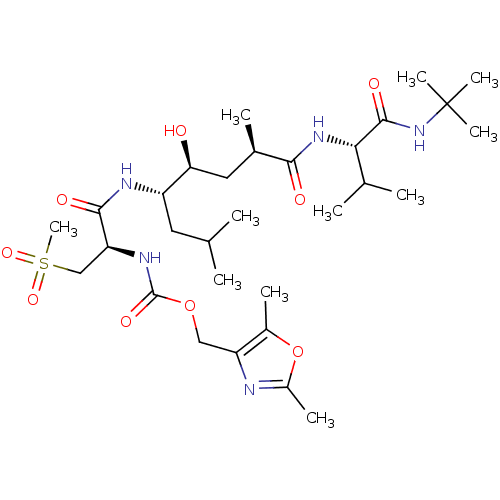
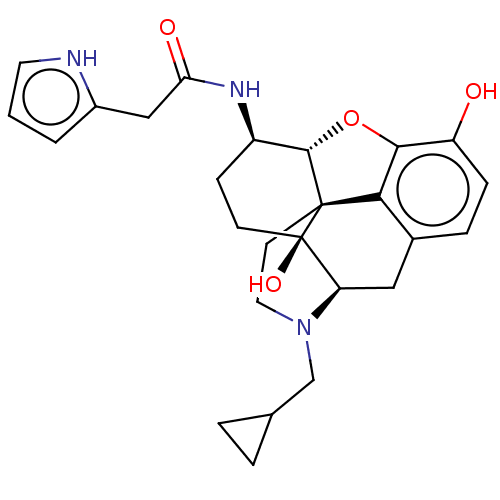
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.00900nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in DrosophilaMore data for this Ligand-Target Pair
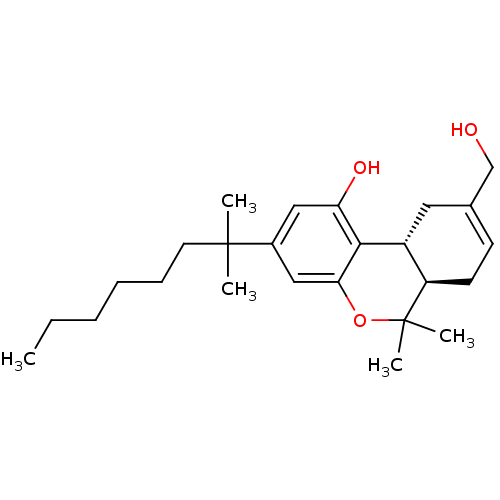
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0100nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in Myzus persicaeMore data for this Ligand-Target Pair
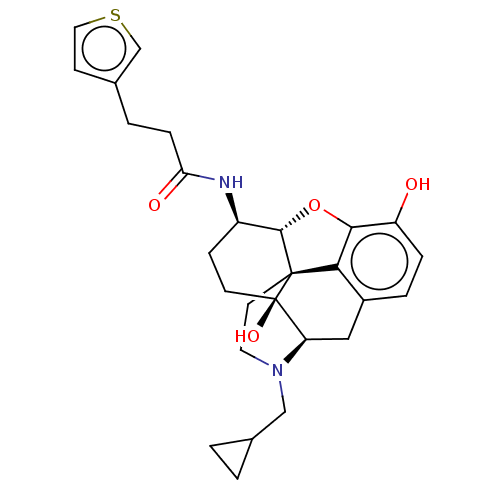
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0110nMAssay Description:Molar concentration required to inhibit 50% of the activating delayed-rectifier K+ current in isolated guinea pig ventricularmyocytesMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0120nM ΔG°: -62.3kJ/mole EC50: 4.60nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0140nM ΔG°: -62.0kJ/mole EC50: 1.20nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
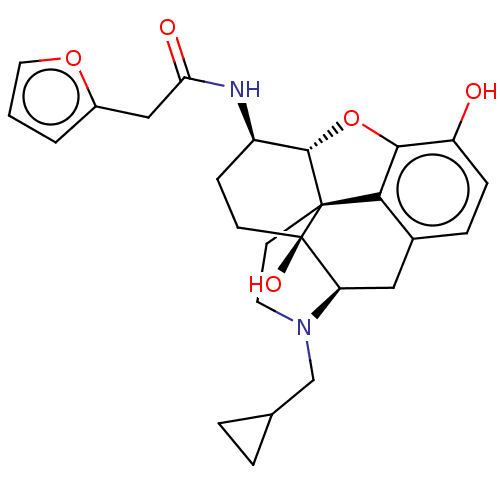
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0150nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in DrosophilaMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0160nM ΔG°: -61.6kJ/mole EC50: 1.60nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0170nM EC50: 14nMAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0230nMAssay Description:Molar concentration required to inhibit 50% of the activating delayed-rectifier K+ current in isolated guinea pig ventricularmyocytesMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0280nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in DrosophilaMore data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0280nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in Myzus persicaeMore data for this Ligand-Target Pair
Affinity DataKi: 0.0300nMAssay Description:Proteolytic activity of SARS-CoV-2 Coronavirus 3CL protease is measured using a continuous fluorescence resonance energy transfer assay. The SARS-CoV...More data for this Ligand-Target Pair
Affinity DataKi: 0.0300nMAssay Description:Proteolytic activity of SARS-CoV-2 Coronavirus 3CL protease is measured using a continuous fluorescence resonance energy transfer assay. The SARS-CoV...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0450nM ΔG°: -59.1kJ/mole EC50: 2nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0510nM ΔG°: -58.7kJ/mole EC50: 15nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0580nM ΔG°: -58.4kJ/mole EC50: 14nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0600nM ΔG°: -58.3kJ/mole EC50: 7nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0700nM EC50: 170nMAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0720nMAssay Description:Molar concentration required to inhibit 50% of the activating delayed-rectifier K+ current in isolated guinea pig ventricularmyocytesMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0770nM ΔG°: -57.7kJ/mole EC50: 20nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0800nM ΔG°: -57.6kJ/mole EC50: 23nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0820nM ΔG°: -57.6kJ/mole EC50: 4nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetNeuronal acetylcholine receptor subunit alpha-4/beta-2(Rattus norvegicus (Rat))
National Institute On Drug Abuse
Curated by ChEMBL
National Institute On Drug Abuse
Curated by ChEMBL
Affinity DataKi: 0.0880nMAssay Description:Binding affinity for nAChR with [3H]-imidacloprid in Myzus persicaeMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0900nM ΔG°: -57.3kJ/mole EC50: 5.5nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Displacement of [3H]CP55940 from CB1 receptor in rat brain membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Displacement of [3H]CP55940 from CB1 receptor in rat brain membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Proteolytic activity of SARS-CoV-2 Coronavirus 3CL protease is measured using a continuous fluorescence resonance energy transfer assay. The SARS-CoV...More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Displacement of [3H]CP-55,940 from mouse CB2 receptor expressed in HEK293 cells by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Displacement of [3H]CP55940 from mouse CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Displacement of [3H]CP55940 from mouse CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.100nM ΔG°: -57.1kJ/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
Affinity DataKi: 0.120nMAssay Description:Binding affinity to recombinant memapsin 2More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.150nM ΔG°: -56.1kJ/mole EC50: 95nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.150nM EC50: 52nMAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
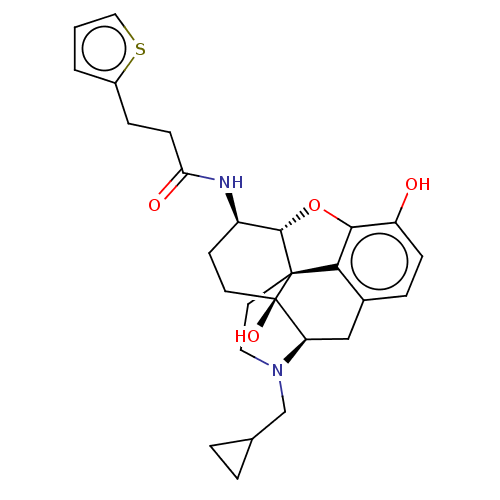
Affinity DataKi: 0.160nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.160nM ΔG°: -55.9kJ/mole EC50: >1.00E+3nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.180nM ΔG°: -55.6kJ/mole EC50: 6.60nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Binding affinity to mouse CB2 receptorMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Binding affinity to mouse CB2 receptorMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]CP55940 from mouse CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]CP55940 from mouse CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]CP55940 from human CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]CP55940 from human CB2 receptor expressed in HEK293 cell membranes by scintillation counting methodMore data for this Ligand-Target Pair
Affinity DataKi: 0.200nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
Affinity DataKi: 0.210nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
Affinity DataKi: 0.210nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
Affinity DataKi: 0.220nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
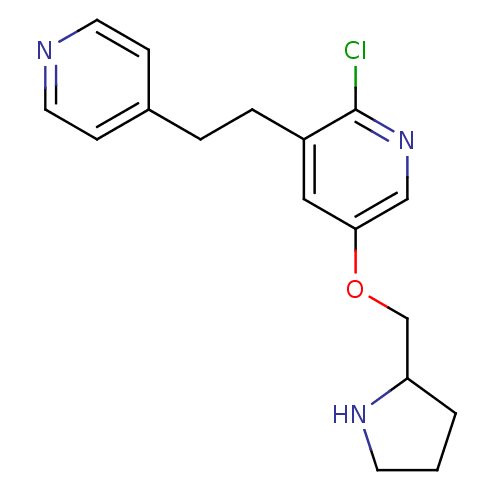
TargetKappa-type opioid receptor(Mus musculus (Mouse))
Virginia Commonwealth University
Curated by ChEMBL
Virginia Commonwealth University
Curated by ChEMBL
Affinity DataKi: 0.220nMAssay Description:Displacement of [3H]-diprenorphine from mouse kappa opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5...More data for this Ligand-Target Pair
Affinity DataKi: 0.230nMAssay Description:Displacement of [3H]-naloxone from mouse mu opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5 hrs by ...More data for this Ligand-Target Pair
TargetKappa-type opioid receptor(Mus musculus (Mouse))
Virginia Commonwealth University
Curated by ChEMBL
Virginia Commonwealth University
Curated by ChEMBL
Affinity DataKi: 0.230nMAssay Description:Displacement of [3H]-diprenorphine from mouse kappa opioid receptor expressed in CHO cell membranes assessed as inhibition constant incubated for 1.5...More data for this Ligand-Target Pair



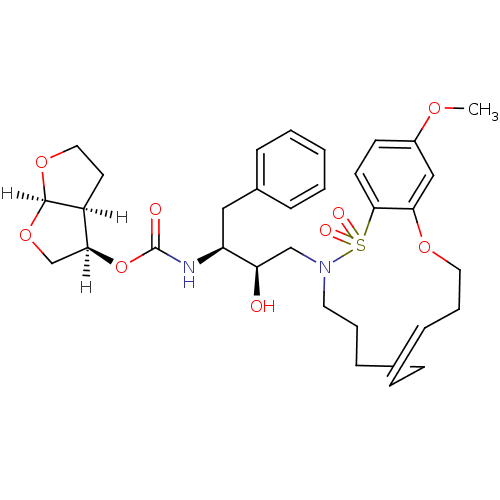
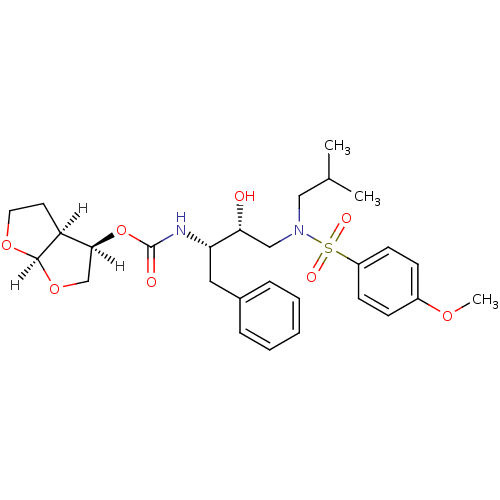
 3D Structure (crystal)
3D Structure (crystal)