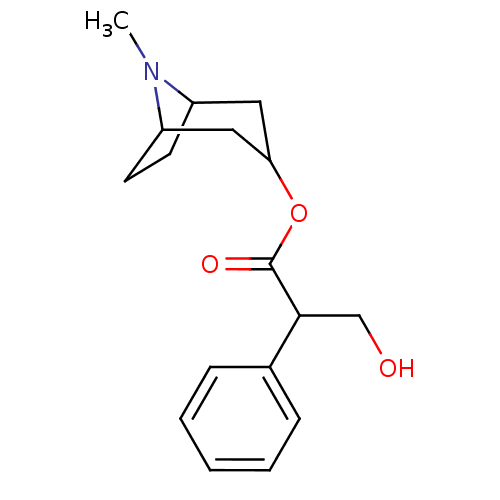
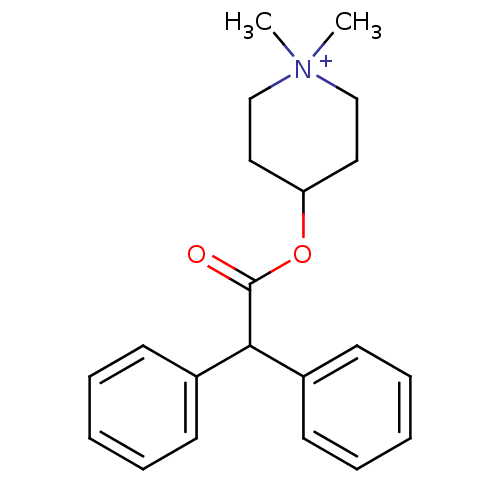
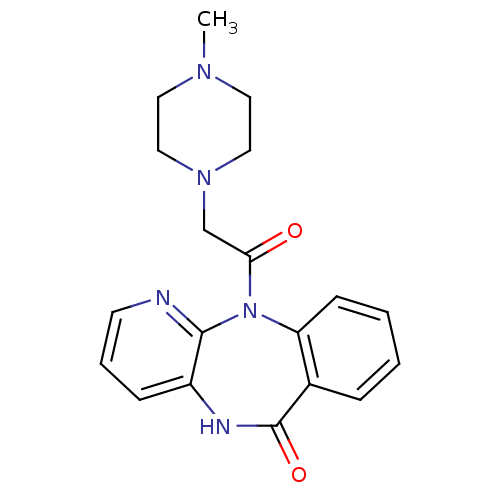
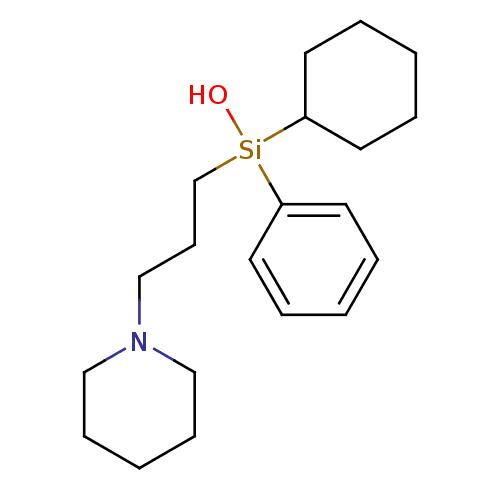
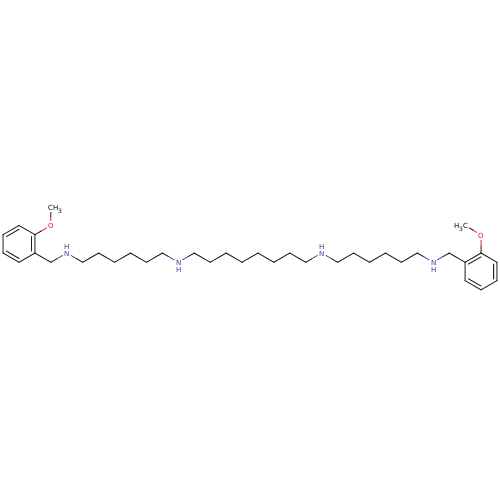
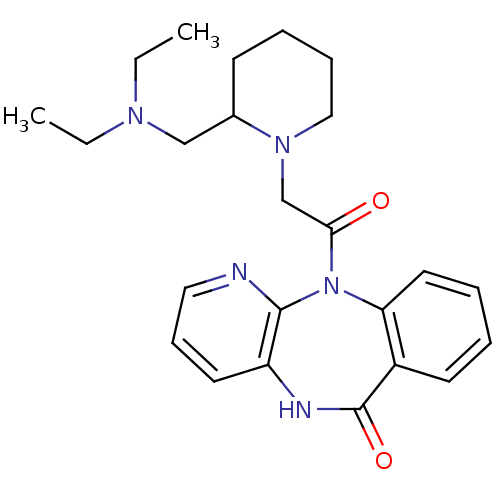
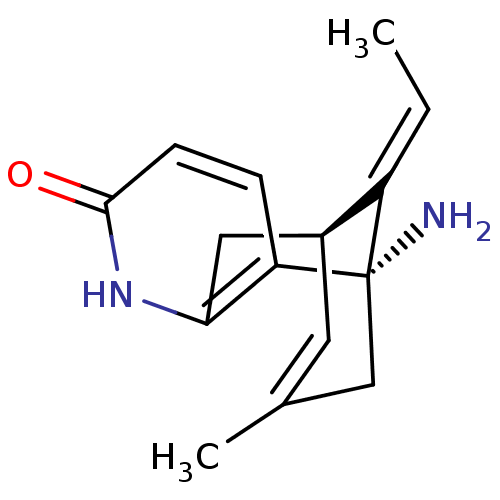
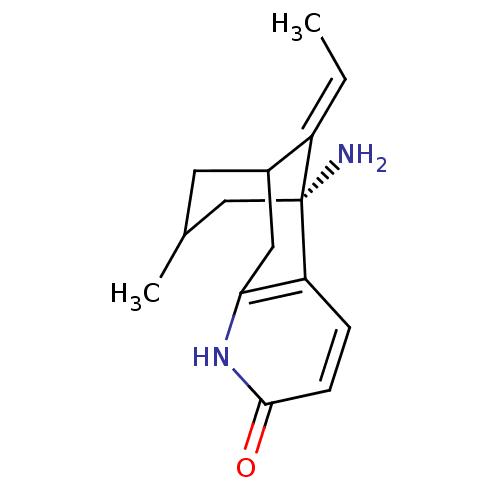
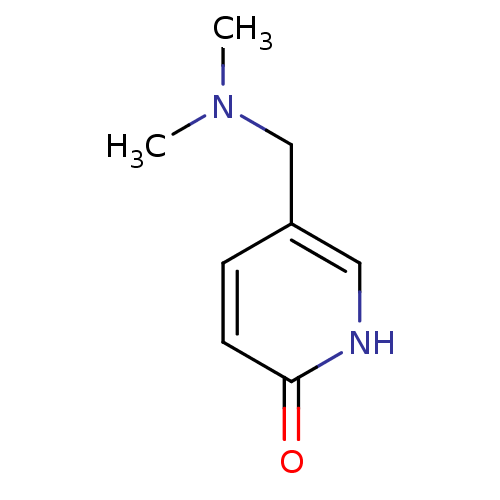
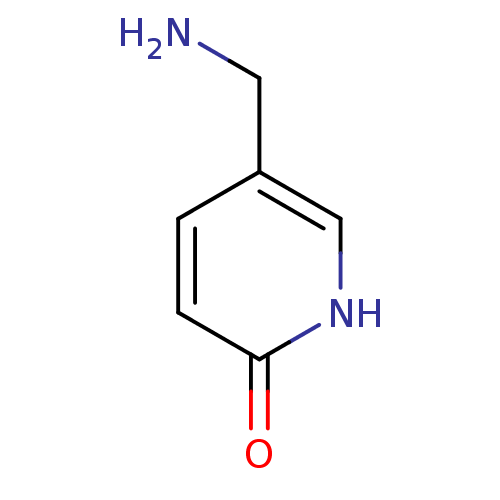
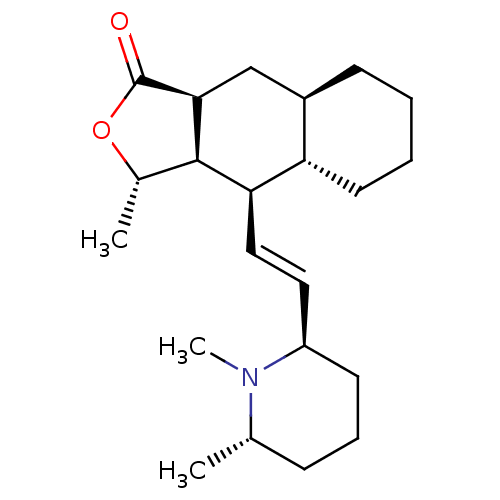
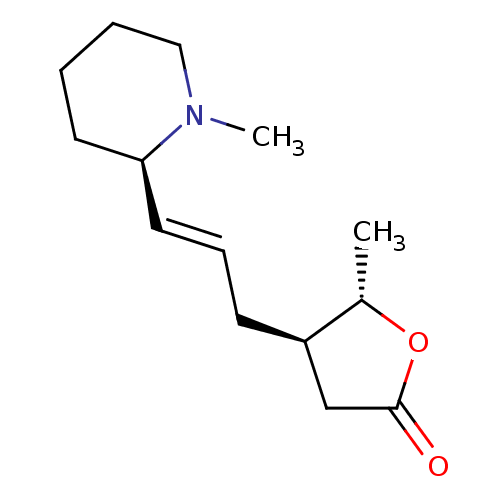
Affinity DataIC50: 44.5nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 45nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 900nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 900nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 9.82E+3nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 3.38E+5nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: >9.00E+5nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: >9.00E+5nMAssay Description:Compound was tested for its ability to inhibit the action of acetylcholinesterase isolated from rat brain cortexMore data for this Ligand-Target Pair
Affinity DataKd: 3nMAssay Description:Dissociation constant for the blocking of cardiac muscarinic M2 receptor was reported.More data for this Ligand-Target Pair
Affinity DataKd: 4.40E+3nMAssay Description:Dissociation constant of the compound for human Muscarinic acetylcholine receptor M2 was determined.More data for this Ligand-Target Pair
Affinity DataKd: 1.21E+3nMAssay Description:Dissociation constant for the blocking of brainstem muscarinic M2 receptor was reported.More data for this Ligand-Target Pair
Affinity DataKd: 11nMAssay Description:Potency of the compound assessed to inhibit cAMP levels in N1E-115 neuroblastoma cells, for blocking oxotremorine-M in functional assay for Muscarini...More data for this Ligand-Target Pair
Affinity DataKd: 6.90nMAssay Description:Dissociation constant for the blocking of heart muscarinic M2 receptor was reported.More data for this Ligand-Target Pair
Affinity DataKd: 4.60nMAssay Description:Dissociation constant for the blocking of brainstem muscarinic M2 receptor was reported.More data for this Ligand-Target Pair
Affinity DataKd: 5.40nMAssay Description:Potency of the compound assessed to inhibit cAMP levels in rat striatum, for blocking oxotremorine-M, in functional assay for Muscarinic acetylcholin...More data for this Ligand-Target Pair
Affinity DataKd: 9.70E+3nMAssay Description:Dissociation constant of the compound for human Muscarinic acetylcholine receptor M1 was determined.More data for this Ligand-Target Pair