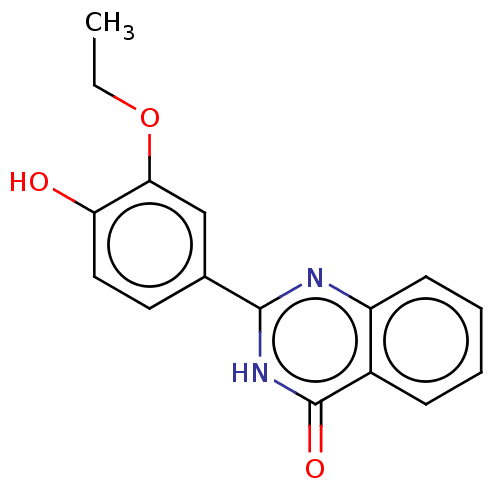
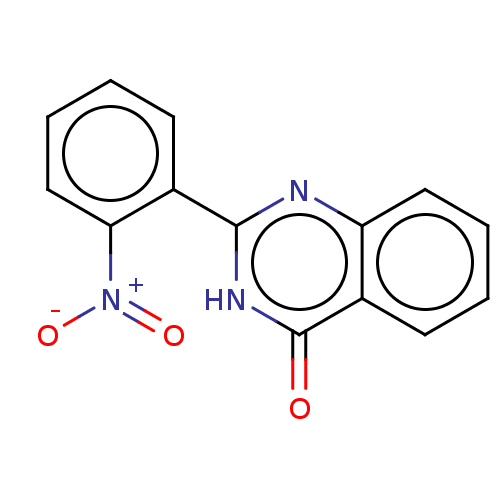
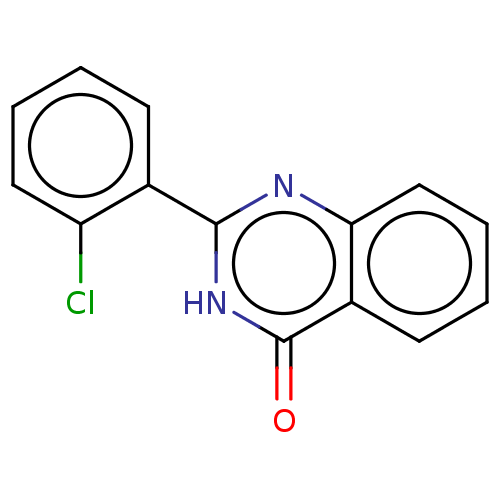
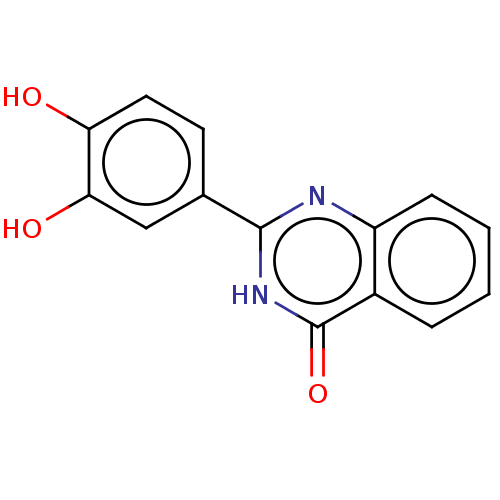
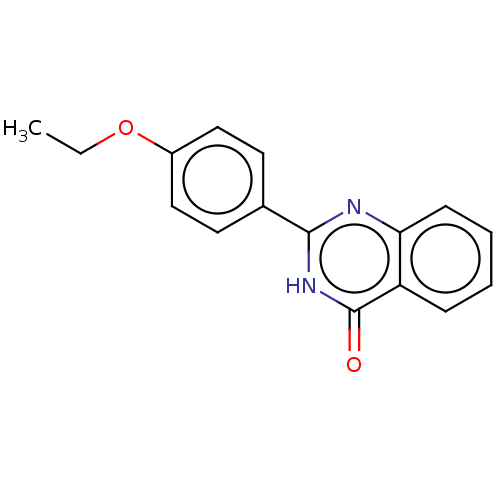
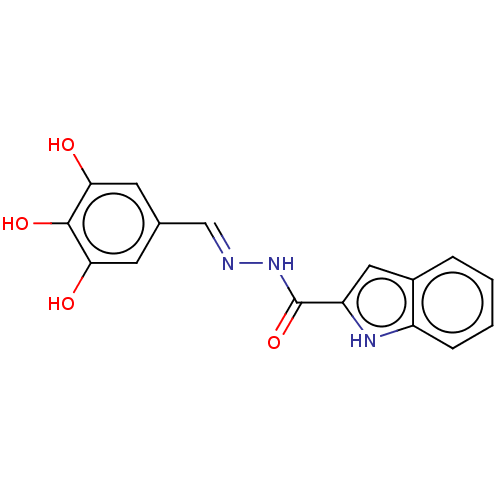
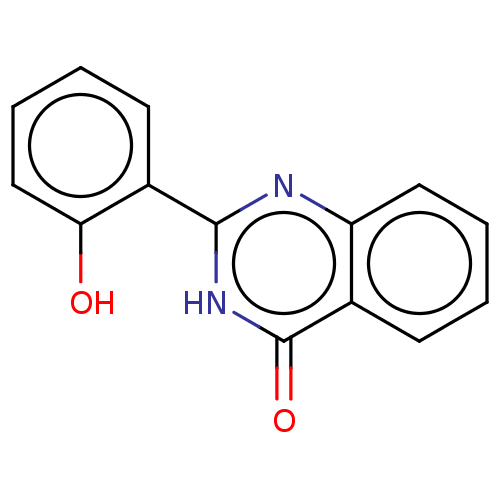
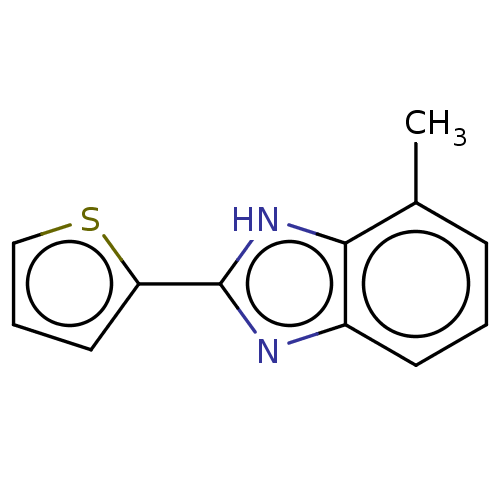
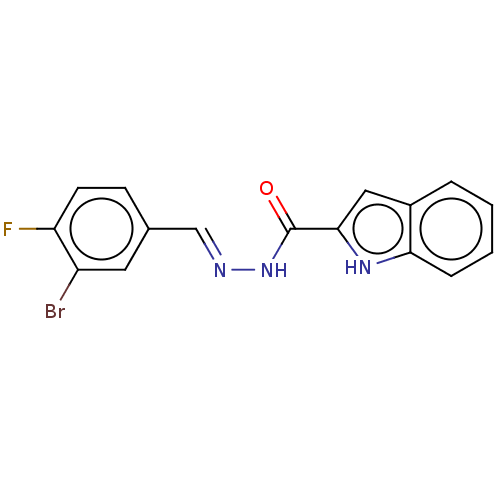
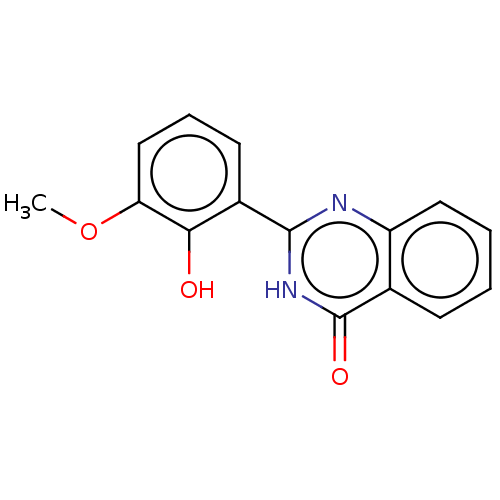
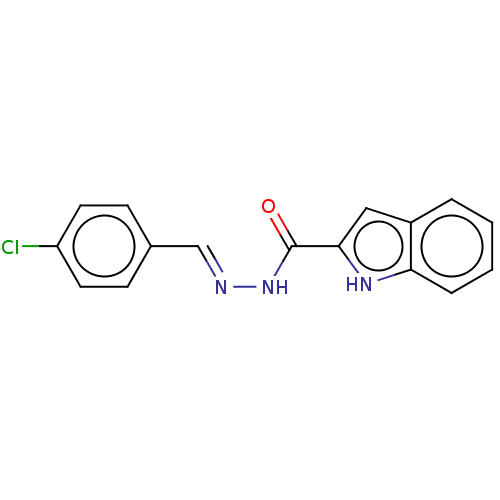
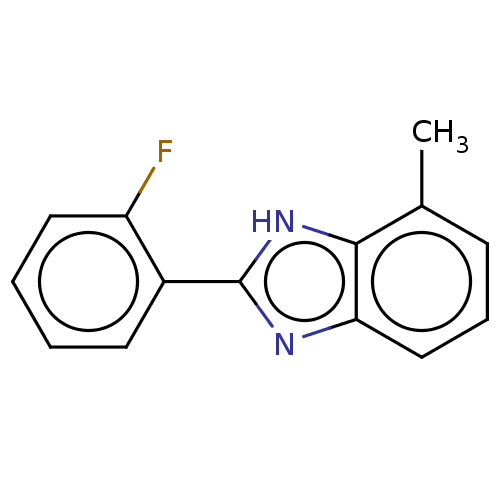
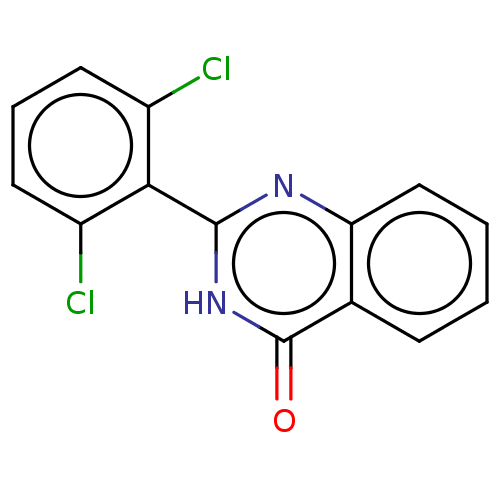
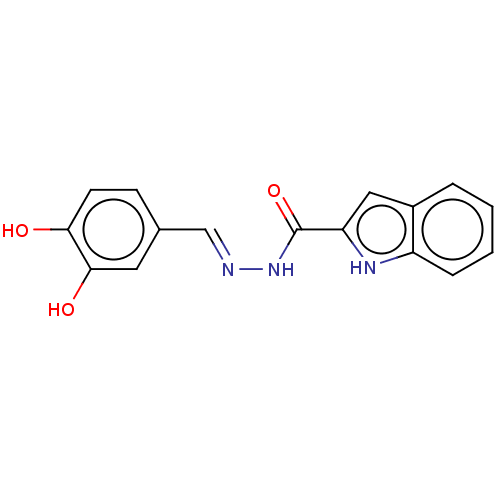
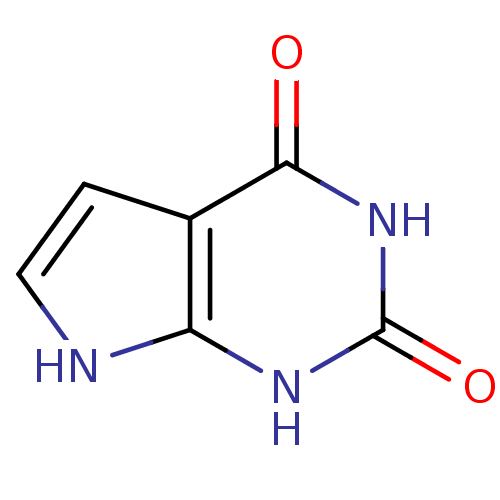
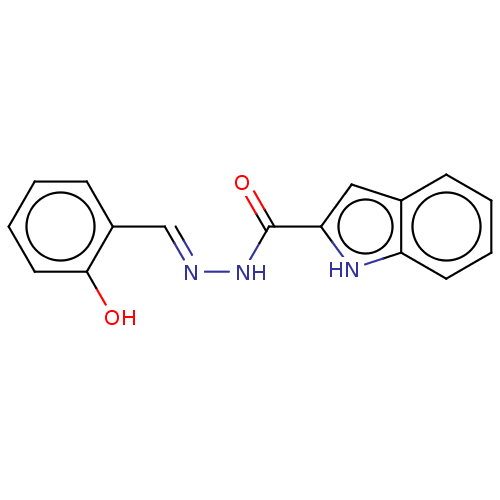
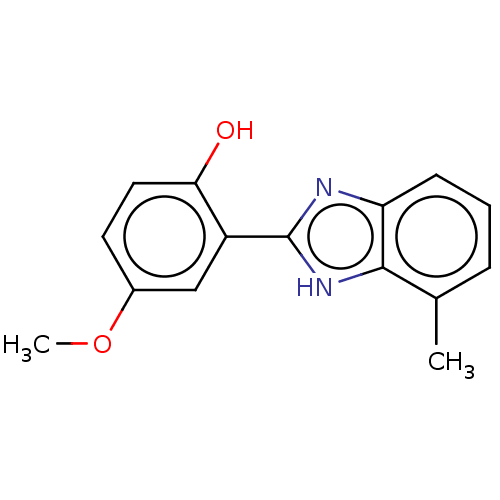
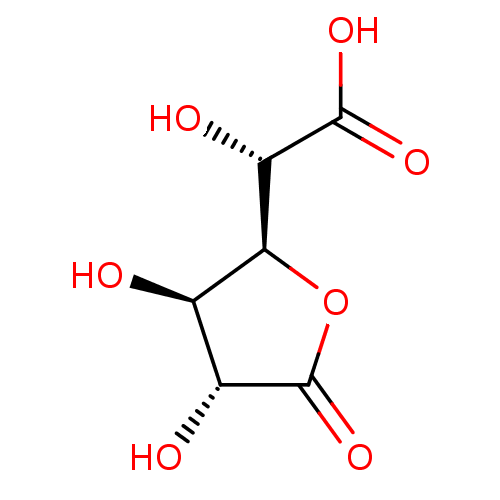
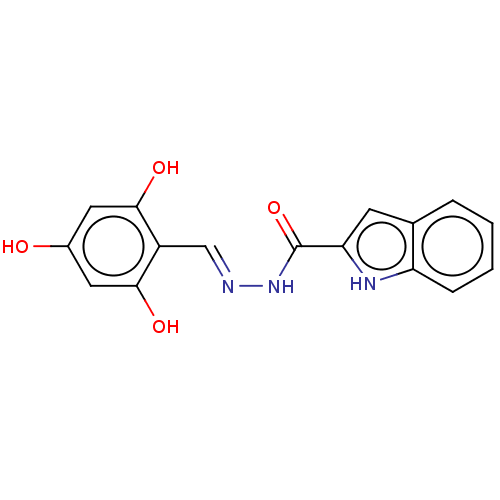
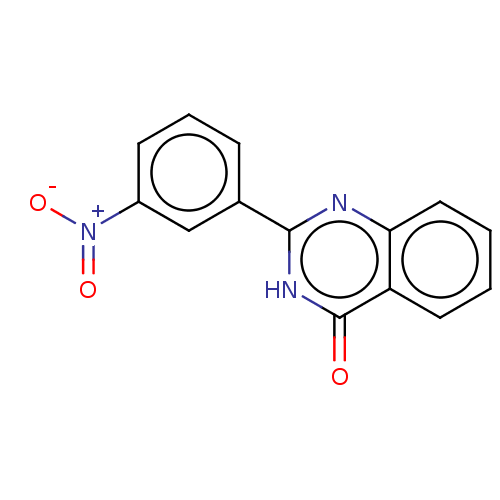
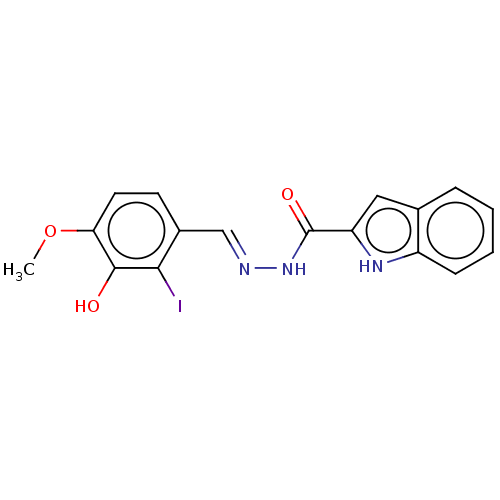
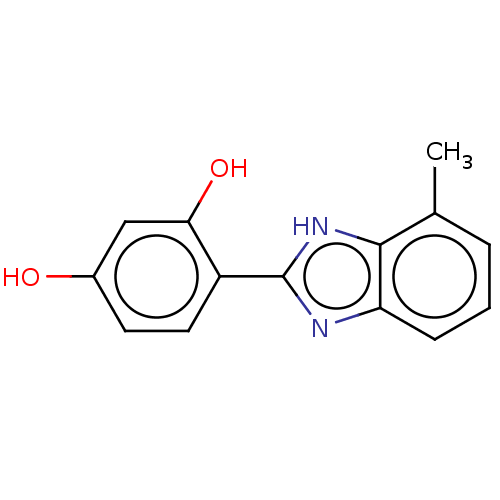
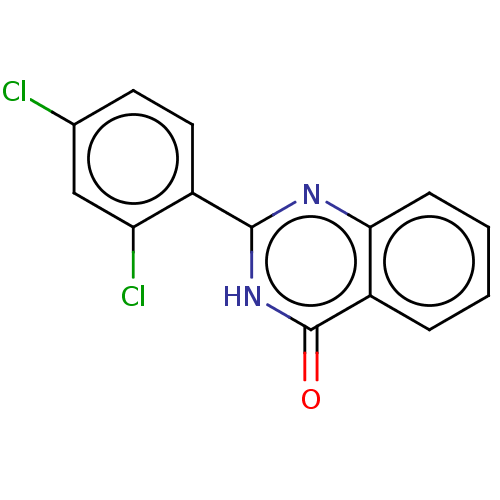
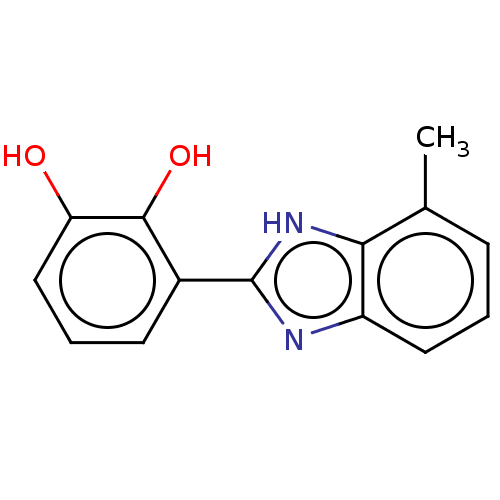
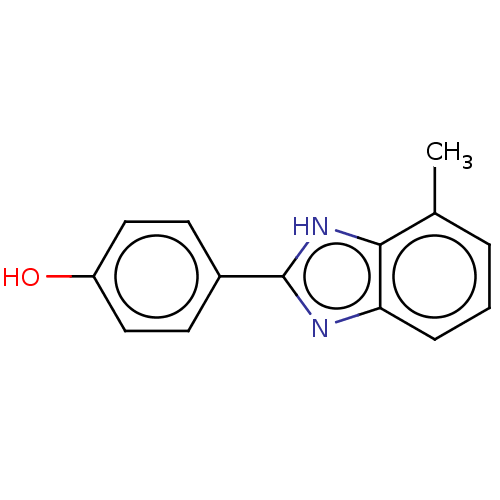
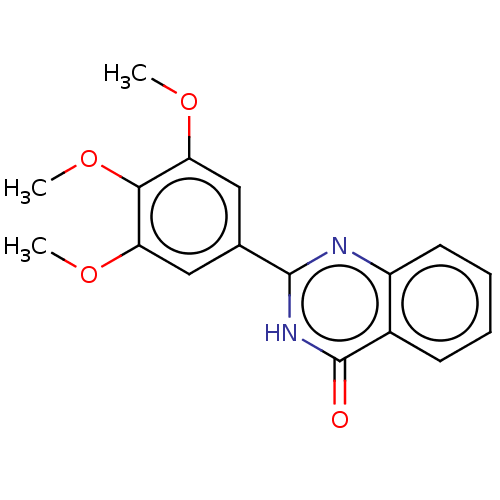
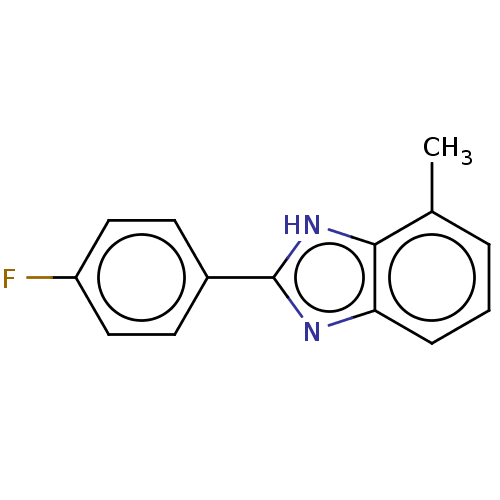
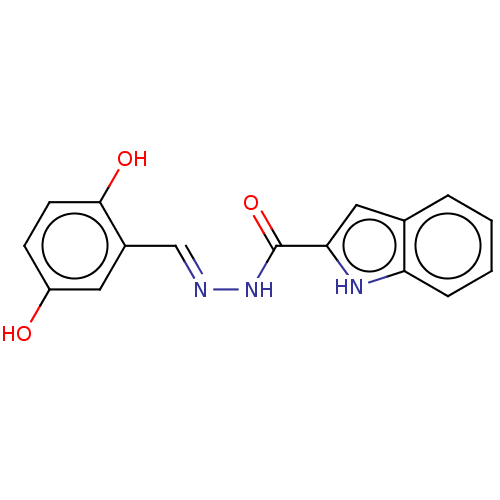
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 165nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 600nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 700nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.10E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.17E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 2.30E+3nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.80E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 3.20E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 5.30E+3nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.50E+3nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 1.61E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.01E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 2.21E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.22E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 2.54E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 2.66E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 2.98E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.09E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 3.21E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 3.44E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.77E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
Affinity DataIC50: 3.98E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.10E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+4nMpH: 7.0Assay Description:TP inhibition assay was performed spectrophotometrically. The method of Bera et al. was followed with slight modifications. Reaction mixture of 200 �...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.26E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.29E+4nMpH: 7.0Assay Description:TP inhibition assay was performed spectrophotometrically. The method of Bera et al. was followed with slight modifications. Reaction mixture of 200 �...More data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.46E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.52E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.58E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.92E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 4.92E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.04E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 5.46E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
Affinity DataIC50: 5.95E+4nMpH: 7.0Assay Description:TP inhibition assay was performed spectrophotometrically. The method of Bera et al. was followed with slight modifications. Reaction mixture of 200 �...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 6.09E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 6.10E+4nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 8.56E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 8.65E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 9.56E+4nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.21E+5nMAssay Description:Inhibition of beta-glucuronidase activity (unknown origin) assessed as p-nitrophenol formation after 30 mins using p-nitrophenyl-beta-D-glucuronide a...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 1.25E+5nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 1.36E+5nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8), solution of enzyme (20 �L), and 20 �L of test sample with 70% DMSO was added to 96-well plate. ...More data for this Ligand-Target Pair
TargetOligo-1,6-glucosidase IMA1(Saccharomyces cerevisiae S288c (Baker's yeast))
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Universiti Teknologi Mara (Uitm), Puncak Alam Campus
Affinity DataIC50: 1.44E+5nMAssay Description:The 135 �L of 50 mM phosphate saline buffer pH (6.8) was added in the 96-well plate and 20 �L of test sample with 70% DMSO added into the wells. The ...More data for this Ligand-Target Pair