Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Prothrombin
Ligand
BDBM50228863
Substrate
n/a
Meas. Tech.
ChEBML_209092
Ki
5.5±n/a nM
Citation
 Zhang, M; Bailey, DL; Bastian, JA; Briggs, SL; Chirgadze, NY; Clawson, DK; Denney, ML; Gifford-Moore, DS; Harper, RW; Johnson, LM; Klimkowski, VJ; Kohn, TJ; Lin, HS; McCowan, JR; Richett, ME; Sall, DJ; Smith, AJ; Smith, GF; Snyder, DW; Takeuchi, K; Utterback, BG; Yan, SC Dibasic benzo[b]thiophene derivatives as a novel class of active site directed thrombin inhibitors: 2. Sidechain optimization and demonstration of in vivo efficacy. Bioorg Med Chem Lett 9:775-80 (1999) [PubMed] Article
Zhang, M; Bailey, DL; Bastian, JA; Briggs, SL; Chirgadze, NY; Clawson, DK; Denney, ML; Gifford-Moore, DS; Harper, RW; Johnson, LM; Klimkowski, VJ; Kohn, TJ; Lin, HS; McCowan, JR; Richett, ME; Sall, DJ; Smith, AJ; Smith, GF; Snyder, DW; Takeuchi, K; Utterback, BG; Yan, SC Dibasic benzo[b]thiophene derivatives as a novel class of active site directed thrombin inhibitors: 2. Sidechain optimization and demonstration of in vivo efficacy. Bioorg Med Chem Lett 9:775-80 (1999) [PubMed] Article More Info.:
Target
Name:
Prothrombin
Synonyms:
Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain
Type:
Protein
Mol. Mass.:
70029.57
Organism:
Homo sapiens (Human)
Description:
P00734
Residue:
622
Sequence:
MAHVRGLQLPGCLALAALCSLVHSQHVFLAPQQARSLLQRVRRANTFLEEVRKGNLERECVEETCSYEEAFEALESSTATDVFWAKYTACETARTPRDKLAACLEGNCAEGLGTNYRGHVNITRSGIECQLWRSRYPHKPEINSTTHPGADLQENFCRNPDSSTTGPWCYTTDPTVRRQECSIPVCGQDQVTVAMTPRSEGSSVNLSPPLEQCVPDRGQQYQGRLAVTTHGLPCLAWASAQAKALSKHQDFNSAVQLVENFCRNPDGDEEGVWCYVAGKPGDFGYCDLNYCEEAVEEETGDGLDEDSDRAIEGRTATSEYQTFFNPRTFGSGEADCGLRPLFEKKSLEDKTERELLESYIDGRIVEGSDAEIGMSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWDKNFTENDLLVRIGKHSRTRYERNIEKISMLEKIYIHPRYNWRENLDRDIALMKLKKPVAFSDYIHPVCLPDRETAASLLQAGYKGRVTGWGNLKETWTANVGKGQPSVLQVVNLPIVERPVCKDSTRIRITDNMFCAGYKPDEGKRGDACEGDSGGPFVMKSPFNNRWYQMGIVSWGEGCDRDGKYGFYTHVFRLKKWIQKVIDQFGE
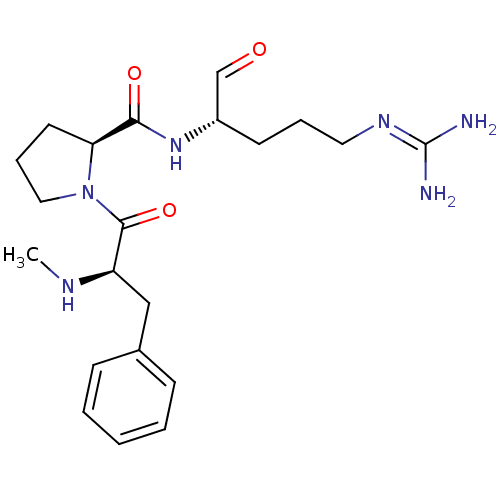
Inhibitor
Name:
BDBM50228863
Synonyms:
(S)-1-((R)-2-Methylamino-3-phenyl-propionyl)-pyrrolidine-2-carboxylic acid ((S)-1-formyl-4-guanidino-butyl)-amide | 1-(2-Methylamino-3-phenyl-propionyl)-pyrrolidine-2-carboxylic acid (1-formyl-4-guanidino-butyl)-amide | 5N-[4-amino(imino)methylamino-1-formyl-(1S)-butyl]-1-[2-methylamino-3-phenyl-(2R)-propanoyl]-(5S)-dihydro-1H-5-pyrrolecarboxamide | CHEMBL273196 | Me-(D-Phe)-Pro-Arg-CHO
Type:
Small organic molecule
Emp. Form.:
C21H32N6O3
Mol. Mass.:
416.5172
SMILES:
[#6]-[#7]-[#6@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@H]-1-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6]=O