Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Dipeptidyl peptidase 9
Ligand
BDBM50323163
Substrate
n/a
Meas. Tech.
ChEMBL_643844 (CHEMBL1211743)
Ki
5900±n/a nM
Citation
 Brigance, RP; Meng, W; Fura, A; Harrity, T; Wang, A; Zahler, R; Kirby, MS; Hamann, LG Synthesis and SAR of azolopyrimidines as potent and selective dipeptidyl peptidase-4 (DPP4) inhibitors for type 2 diabetes. Bioorg Med Chem Lett 20:4395-8 (2010) [PubMed] Article
Brigance, RP; Meng, W; Fura, A; Harrity, T; Wang, A; Zahler, R; Kirby, MS; Hamann, LG Synthesis and SAR of azolopyrimidines as potent and selective dipeptidyl peptidase-4 (DPP4) inhibitors for type 2 diabetes. Bioorg Med Chem Lett 20:4395-8 (2010) [PubMed] Article More Info.:
Target
Name:
Dipeptidyl peptidase 9
Synonyms:
DPP9 | DPP9_HUMAN | DPRP-2 | DPRP2 | Dipeptidyl peptidase 9 (DDP9) | Dipeptidyl peptidase 9 (DPP-9) | Dipeptidyl peptidase 9 (DPP9) | Dipeptidyl peptidase IV-related protein 2 | Dipeptidyl peptidase IX | Dipeptidyl peptidase IX (DDP-IX) | Dipeptidyl peptidase-like protein 9
Type:
Enzyme
Mol. Mass.:
98260.70
Organism:
Homo sapiens (Human)
Description:
Q86TI2
Residue:
863
Sequence:
MATTGTPTADRGDAAATDDPAARFQVQKHSWDGLRSIIHGSRKYSGLIVNKAPHDFQFVQKTDESGPHSHRLYYLGMPYGSRENSLLYSEIPKKVRKEALLLLSWKQMLDHFQATPHHGVYSREEELLRERKRLGVFGITSYDFHSESGLFLFQASNSLFHCRDGGKNGFMVSPMKPLEIKTQCSGPRMDPKICPADPAFFSFINNSDLWVANIETGEERRLTFCHQGLSNVLDDPKSAGVATFVIQEEFDRFTGYWWCPTASWEGSEGLKTLRILYEEVDESEVEVIHVPSPALEERKTDSYRYPRTGSKNPKIALKLAEFQTDSQGKIVSTQEKELVQPFSSLFPKVEYIARAGWTRDGKYAWAMFLDRPQQWLQLVLLPPALFIPSTENEEQRLASARAVPRNVQPYVVYEEVTNVWINVHDIFYPFPQSEGEDELCFLRANECKTGFCHLYKVTAVLKSQGYDWSEPFSPGEDEFKCPIKEEIALTSGEWEVLARHGSKIWVNEETKLVYFQGTKDTPLEHHLYVVSYEAAGEIVRLTTPGFSHSCSMSQNFDMFVSHYSSVSTPPCVHVYKLSGPDDDPLHKQPRFWASMMEAASCPPDYVPPEIFHFHTRSDVRLYGMIYKPHALQPGKKHPTVLFVYGGPQVQLVNNSFKGIKYLRLNTLASLGYAVVVIDGRGSCQRGLRFEGALKNQMGQVEIEDQVEGLQFVAEKYGFIDLSRVAIHGWSYGGFLSLMGLIHKPQVFKVAIAGAPVTVWMAYDTGYTERYMDVPENNQHGYEAGSVALHVEKLPNEPNRLLILHGFLDENVHFFHTNFLVSQLIRAGKPYQLQIYPNERHSIRCPESGEHYEVTLLHFLQEYL
Inhibitor
Name:
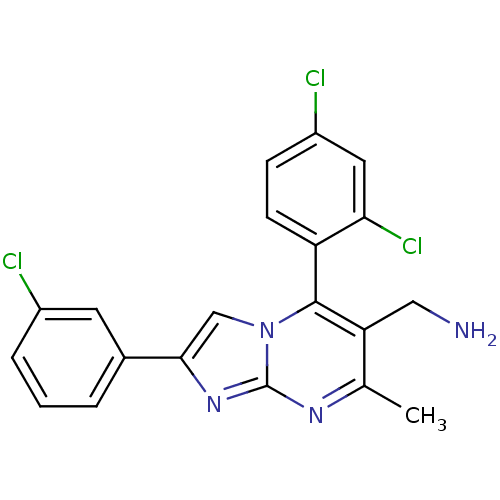
BDBM50323163
Synonyms:
(2-(3-chlorophenyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidin-6-yl)methanamine | CHEMBL1208841
Type:
Small organic molecule
Emp. Form.:
C20H15Cl3N4
Mol. Mass.:
417.719
SMILES:
Cc1nc2nc(cn2c(c1CN)-c1ccc(Cl)cc1Cl)-c1cccc(Cl)c1 |(2.01,-11.55,;.68,-12.32,;-.66,-11.55,;-1.99,-12.33,;-3.47,-11.86,;-4.36,-13.12,;-3.45,-14.36,;-1.98,-13.87,;-.65,-14.63,;.68,-13.86,;2.02,-14.63,;3.35,-13.86,;-.64,-16.17,;-1.97,-16.94,;-1.97,-18.48,;-.64,-19.25,;-.64,-20.79,;.7,-18.48,;.69,-16.93,;2.02,-16.16,;-5.9,-13.13,;-6.68,-11.8,;-8.22,-11.81,;-8.98,-13.15,;-8.19,-14.48,;-8.95,-15.82,;-6.66,-14.46,)|