Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
E3 ubiquitin-protein ligase Mdm2
Ligand
BDBM50339370
Substrate
n/a
Meas. Tech.
ChEMBL_736488 (CHEMBL1694433)
IC50
390±n/a nM
Citation
 Hardcastle, IR; Liu, J; Valeur, E; Watson, A; Ahmed, SU; Blackburn, TJ; Bennaceur, K; Clegg, W; Drummond, C; Endicott, JA; Golding, BT; Griffin, RJ; Gruber, J; Haggerty, K; Harrington, RW; Hutton, C; Kemp, S; Lu, X; McDonnell, JM; Newell, DR; Noble, ME; Payne, SL; Revill, CH; Riedinger, C; Xu, Q; Lunec, J Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J Med Chem 54:1233-43 (2011) [PubMed] Article
Hardcastle, IR; Liu, J; Valeur, E; Watson, A; Ahmed, SU; Blackburn, TJ; Bennaceur, K; Clegg, W; Drummond, C; Endicott, JA; Golding, BT; Griffin, RJ; Gruber, J; Haggerty, K; Harrington, RW; Hutton, C; Kemp, S; Lu, X; McDonnell, JM; Newell, DR; Noble, ME; Payne, SL; Revill, CH; Riedinger, C; Xu, Q; Lunec, J Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J Med Chem 54:1233-43 (2011) [PubMed] Article More Info.:
Target
Name:
E3 ubiquitin-protein ligase Mdm2
Synonyms:
Double minute 2 protein | Double minute 2 protein (HDM2) | E3 ubiquitin-protein ligase Mdm2 (p53-binding protein Mdm2) | Hdm2 | Human Double Minute 2 (HDM2) | MDM2 | MDM2-MDMX | MDM2_HUMAN | p53-Binding Protein MDM2 | p53-binding protein
Type:
Oncoprotein
Mol. Mass.:
55196.54
Organism:
Homo sapiens (Human)
Description:
Q00987
Residue:
491
Sequence:
MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEVYQVTVYQAGESDTDSFEEDPEISLADYWKCTSCNEMNPPLPSHCNRCWALRENWLPEDKGKDKGEISEKAKLENSTQAEEGFDVPDCKKTIVNDSRESCVEENDDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQDKEESVESSLPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQPIQMIVLTYFP
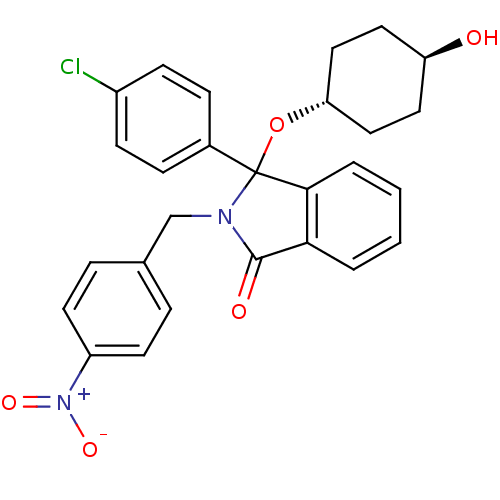
Inhibitor
Name:
BDBM50339370
Synonyms:
(+/-)-trans-3-(4-Chlorophenyl)-3-(4-hydroxycyclohexyloxy)-2-(4-nitrobenzyl)-2,3-dihydroisoindol-1-one | CHEMBL1688275
Type:
Small organic molecule
Emp. Form.:
C27H25ClN2O5
Mol. Mass.:
492.951
SMILES:
O[C@H]1CC[C@@H](CC1)OC1(N(Cc2ccc(cc2)[N+]([O-])=O)C(=O)c2ccccc12)c1ccc(Cl)cc1 |r,wU:4.7,wD:1.0,(-9.71,-36.66,;-8.4,-37.46,;-8.44,-39,;-7.12,-39.8,;-5.79,-39.06,;-5.74,-37.53,;-7.05,-36.73,;-4.48,-39.85,;-4.08,-41.35,;-3.16,-42.6,;-1.62,-42.6,;-.85,-43.93,;-1.62,-45.27,;-.86,-46.6,;.69,-46.6,;1.46,-45.26,;.68,-43.93,;1.47,-47.94,;.7,-49.28,;3.01,-47.94,;-4.08,-43.86,;-3.6,-45.32,;-5.55,-43.38,;-6.88,-44.15,;-8.22,-43.38,;-8.22,-41.83,;-6.89,-41.06,;-5.55,-41.82,;-2.76,-40.55,;-1.41,-41.31,;-.09,-40.52,;-.11,-38.97,;1.21,-38.18,;-1.47,-38.23,;-2.79,-39.02,)|