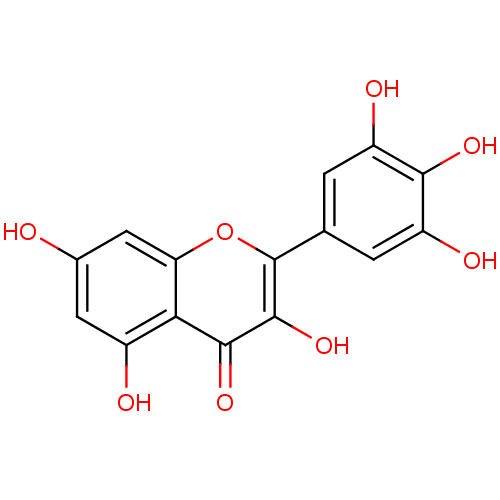
BDBM15236 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)-4H-chromen-4-one::3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromen-4-one::CHEMBL164::Cannabiscetin::Myricetin::Myricetin (20)::Myricetin (Myr)::cid_5281672
SMILES c1c(cc(c(c1O)O)O)C2=C(C(=O)c3c(cc(cc3O2)O)O)O
InChI Key InChIKey=IKMDFBPHZNJCSN-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 134 hits for monomerid = 15236
Found 134 hits for monomerid = 15236
Affinity DataKd: 0.25nMAssay Description:Binding affinity to MBNL1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataEC50: 0.510nMAssay Description:Activation of recombinant PKM2 (unknown origin) Phe26, Tyr390, Glu397, Asp354, Leu353 residuesMore data for this Ligand-Target Pair
Affinity DataIC50: 12nMAssay Description:Inhibition of AChE (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 27nMAssay Description:Inhibition of CYP1B1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 27nMAssay Description:Inhibition of CYP1B1 EROD activity assessed as inhibition of deethylation of 7-ethoxyresorufin to resorufinMore data for this Ligand-Target Pair
Affinity DataKd: 170nMAssay Description:Binding affinity to PI3Kalpha (unknown origin) assessed as dissociation constantMore data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+3nM Kd: 170nMpH: 7.2 T: 2°CAssay Description:Binding was detected as a change in the intrinsic tryptophan fluorescence of the PI3K upon the addition of inhibitor. The inhibitor was incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:Inhibition of SARS-CoV-2 Main proteaseMore data for this Ligand-Target Pair
Affinity DataIC50: 220nMAssay Description:Inhibition of alpha-synuclein fibril formation (unknown origin) incubated for 24 hrs to 7 days by thioflavin S based fluorescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 220nMAssay Description:Primary assay principle based on quenched FRET peptide substrate of SARS-CoV-2 3CL-Pro (lhs). Inhibiting compounds reduce fluorescence signal relativ...More data for this Ligand-Target Pair
Affinity DataIC50: 260nMAssay Description:Primary assay principle based on quenched FRET peptide substrate of SARS-CoV-2 3CL-Pro (lhs). Inhibiting compounds reduce fluorescence signal relativ...More data for this Ligand-Target Pair
TargetDNA-(apurinic or apyrimidinic site) endonuclease(Human)
Russian Academy of Sciences
Curated by ChEMBL
Russian Academy of Sciences
Curated by ChEMBL
Affinity DataIC50: 320nMAssay Description:Inhibition of human APE1 preincubated for 15 mins followed by substrate addition by HTS assayMore data for this Ligand-Target Pair
Affinity DataKi: 370nMAssay Description:Inhibition of CYP1A1 EROD activity assessed as inhibition of deethylation of 7-ethoxyresorufin to resorufinMore data for this Ligand-Target Pair
Affinity DataIC50: 390nMAssay Description:Inhibition of Homo sapiens (human) DNA topoisomerase 2More data for this Ligand-Target Pair
TargetEnoyl-acyl-carrier protein reductase(malaria parasite P. falciparum)
University of Zurich
Curated by ChEMBL
University of Zurich
Curated by ChEMBL
Affinity DataIC50: 415nMAssay Description:Inhibition of human ENPP1More data for this Ligand-Target Pair
Affinity DataIC50: 560nMAssay Description:Inhibition of recombinant human His-tagged glyoxalase 1 expressed in Escherichia coli BL21 assessed as formation of S-D-lactoylglutathione after 5 mi...More data for this Ligand-Target Pair
Affinity DataIC50: 560nMAssay Description:Inhibition of human recombinant His-tagged Glyoxalase 1 expressed in Sf21-Baculovirus systemMore data for this Ligand-Target Pair
Affinity DataIC50: 600nMAssay Description:Inhibition of full length SARS-CoV-2 3CL protease expressed in Escherichia coli BL21 (DE3) using MCA-AVLQSGFR-Lys(Dnp)-Lys-NH2 as substrate preincuba...More data for this Ligand-Target Pair
Affinity DataIC50: 600nMAssay Description:Tested for inhibition of HIV-1 integrase, under 1 uM for the strand transferMore data for this Ligand-Target Pair
Affinity DataIC50: 630nMAssay Description:Inhibition of SARS-CoV-2 3CLp using fluorescent substrate by FRET assayMore data for this Ligand-Target Pair
Affinity DataIC50: 630nMAssay Description:Inhibition of SARS-CoV-2 MPro using Dabcyl-KTSAVLQSGFRKME-Edans as substrate preincubated for 30 mins followed by substrate addition measured after 1...More data for this Ligand-Target Pair
Affinity DataIC50: 630nMAssay Description:Inhibition of SARS CoV-2 main proteaseMore data for this Ligand-Target Pair
TargetInositol hexakisphosphate kinase 2(Human)
National Institute of Environmental Health Sciences
Curated by ChEMBL
National Institute of Environmental Health Sciences
Curated by ChEMBL
Affinity DataIC50: 700nMAssay Description:Inhibition of human IP6K2 using insP6 as substrate preincubated for 15 mins followed by substrate and measured after 30 mins by TR-FRET assayMore data for this Ligand-Target Pair
TargetPC4 and SFRS1-interacting protein(Human)
The Scripps Research Institute Molecular Screening Center
Curated by PubChem BioAssay
The Scripps Research Institute Molecular Screening Center
Curated by PubChem BioAssay
Affinity DataIC50: 758nMAssay Description:Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: Ohio State University Assay Provider: Mam...More data for this Ligand-Target Pair
TargetSerine/threonine-protein kinase pim-1(Human)
Loma Linda University School of Medicine
Curated by ChEMBL
Loma Linda University School of Medicine
Curated by ChEMBL
Affinity DataIC50: 776nMAssay Description:Inhibition of PIM1 kinaseMore data for this Ligand-Target Pair
TargetSerine/threonine-protein kinase pim-1(Human)
Loma Linda University School of Medicine
Curated by ChEMBL
Loma Linda University School of Medicine
Curated by ChEMBL
Affinity DataIC50: 780nMpH: 7.0 T: 2°CAssay Description:The 96-well flat-bottomed plates were coated with recombinant GST-BAD. After the plates were blocked, the reaction buffer containing test compound an...More data for this Ligand-Target Pair
TargetSerine/threonine-protein kinase pim-1(Human)
Loma Linda University School of Medicine
Curated by ChEMBL
Loma Linda University School of Medicine
Curated by ChEMBL
Affinity DataIC50: 780nMAssay Description:Inhibition of recombinant PIM1 (unknown origin) expressed in Escherichia coli after 60 mins by ELISAMore data for this Ligand-Target Pair
Affinity DataKi: 800nMAssay Description:Inhibition of Zika virus Asian/8375 NS2B (48 to 100 residues)-NS3 (14 to 185 residues) expressed in Escherichia coli BL21 (DE3) Star cells preincubat...More data for this Ligand-Target Pair
TargetProtein E6(Human papillomavirus type 16)
Loma Linda University School of Medicine
Curated by ChEMBL
Loma Linda University School of Medicine
Curated by ChEMBL
Affinity DataIC50: 850nMAssay Description:Inhibition of GST-tagged Human papillomavirus 16 protein E6 interaction with His-tagged human caspase 8 expressed in Escherichia coli after 1 hr incu...More data for this Ligand-Target Pair
Affinity DataIC50: 900nMAssay Description:Inhibition of human recombinant CK2 expressed in Escherichia coli using RRRADDSDDDDD as substrate after 10 mins in presence of [gamma-32P]ATPMore data for this Ligand-Target Pair
Affinity DataIC50: 920nMAssay Description:Inhibition of rat liver CK2 phosphorylation using RRRADDSDDDDD as substrate in presence of [32p]-ATPMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:Inhibition of ELAV3 (unknown origin)-artificial ARE complex formation after 30 mins in the presence of biotin-labeled RNA probe by chemiluminescence ...More data for this Ligand-Target Pair
TargetInositol polyphosphate multikinase(Human)
National Institute of Environmental Health Sciences
Curated by ChEMBL
National Institute of Environmental Health Sciences
Curated by ChEMBL
Affinity DataIC50: 1.10E+3nMAssay Description:Inhibition of human IPMK using insP3 as substrate preincubated for 15 mins followed by substrate and measured after 30 mins by TR-FRET assayMore data for this Ligand-Target Pair
TargetPC4 and SFRS1-interacting protein(Human)
The Scripps Research Institute Molecular Screening Center
Curated by PubChem BioAssay
The Scripps Research Institute Molecular Screening Center
Curated by PubChem BioAssay
Affinity DataIC50: 1.14E+3nMAssay Description:Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: Ohio State University Assay Provider: Mam...More data for this Ligand-Target Pair
Affinity DataKi: 1.20E+3nMAssay Description:Noncompetitive inhibition of Leishmania amazonensis recombinant arginase expressed in Escherichia coli Rosetta (DE3) pLysS using L-arginine as substr...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:Inhibition of human recombinant brain tau protein (412 amino acid residues) filament assembly expressed in Escherichia coli BL21(DE3) by electron mic...More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+3nMAssay Description:Inhibition of Zika virus Asian/8375 NS2B (48 to 100 residues)-NS3 (14 to 185 residues) expressed in Escherichia coli BL21 (DE3) Star cells preincubat...More data for this Ligand-Target Pair
TargetPancreatic triacylglycerol lipase(Human)
Shanghai University of Traditional Chinese Medicine
Curated by ChEMBL
Shanghai University of Traditional Chinese Medicine
Curated by ChEMBL
Affinity DataIC50: 1.34E+3nMAssay Description:Negative allosteric modulator activity at human Pancreatic lipase using DDAO-ol as fluorescent substrate preincubated for 3 mins followed by substrat...More data for this Ligand-Target Pair
Target1-deoxy-D-xylulose 5-phosphate reductoisomerase(Escherichia coli (strain K12))
University of Strasburg
University of Strasburg
Affinity DataIC50: 1.50E+3nMAssay Description:H-DXR was pre-incubated during 2 min in the presence of the inhibitors at different concentrations and DXP (480 µM). NADPH (160 µM final concentratio...More data for this Ligand-Target Pair
Affinity DataIC50: 1.64E+3nMT: 2°CAssay Description:The p38alpha reaction was carried out by using kinase (12ng per well), ATP (100uM) and incubated for 60 min at 37 C. For the JNK3 assay, kinase (10n...More data for this Ligand-Target Pair
Affinity DataIC50: 1.69E+3nMAssay Description:Inhibition of recombinant FLT3 (unknown origin) by TR-FRET assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.80E+3nMAssay Description:Inhibition of Fujinami sarcoma virus pp130fpsMore data for this Ligand-Target Pair
Target3-hydroxyacyl-[acyl-carrier-protein] dehydratase(malaria parasite P. falciparum)
University of Zurich
Curated by ChEMBL
University of Zurich
Curated by ChEMBL
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of FabZMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of HIV-1 integrase, under 1 uM for the 3''-preprocessingMore data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+3nMAssay Description:Inhibition of Leishmania amazonensis recombinant arginase expressed in Escherichia coli Rosetta (DE3) pLysS using L-arginine as substrate incubated f...More data for this Ligand-Target Pair
Affinity DataIC50: 2.12E+3nMAssay Description:Desensitization of GPR35 receptor in human HT-29 cells assessed as inhibition of zaprinast-induced dynamic mass redistribution after 10 minsMore data for this Ligand-Target Pair
Affinity DataIC50: 2.24E+3nMT: 2°CAssay Description:The p38alpha reaction was carried out by using kinase (12ng per well), ATP (100uM) and incubated for 60 min at 37 C. For the JNK3 assay, kinase (10n...More data for this Ligand-Target Pair
Affinity DataIC50: 2.38E+3nMAssay Description:Inhibition of xanthine oxidase assessed as decrease in uric acid production by spectrophotometryMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)