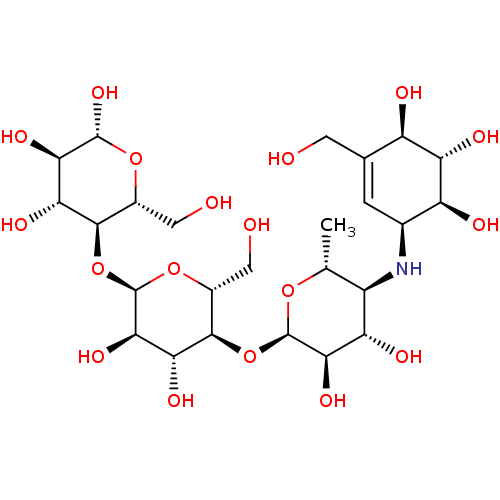
BDBM50333465 (2R,3R,4R,5R,6R)-5-((2R,3R,4R,5S,6R)-5-((2R,3R,4S,5S,6R)-3,4-dihydroxy-6-methyl-5-((1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-enylamino)-tetrahydro-2H-pyran-2-yloxy)-3,4-dihydroxy-6-(hydroxymethyl)-tetrahydro-2H-pyran-2-yloxy)-6-(hydroxymethyl)-tetrahydro-2H-pyran-2,3,4-triol::(2R,3R,4R,5S,6R)-5-((2R,3R,4R,5S,6R)-5-((2R,3R,4S,5S,6R)-3,4-dihydroxy-6-methyl-5-((1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-enylamino)-tetrahydro-2H-pyran-2-yloxy)-3,4-dihydroxy-6-(hydroxymethyl)-tetrahydro-2H-pyran-2-yloxy)-6-(hydroxymethyl)-tetrahydro-2H-pyran-2,3,4-triol::(2R,3R,4R,5S,6R)-5-((2R,3R,4R,5S,6R)-5-((2R,3R,4S,5S,6R)-3,4-dihydroxy-6-methyl-5-((1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-enylamino)-tetrahydro-2H-pyran-2-yloxy)-3,4-dihydroxy-6-(hydroxymethyl)-tetrahydro-2H-pyran-2-yloxy)-6-(hydroxymethyl)-tetrahydro-2H-pyran-2,3,4-triol::(3R,4R,5S,6R)-5-((2R,3R,4R,5S,6R)-5-((2R,3R,4S,5S,6R)-3,4-dihydroxy-6-methyl-5-((1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-enylamino)-tetrahydro-2H-pyran-2-yloxy)-3,4-dihydroxy-6-(hydroxymethyl)-tetrahydro-2H-pyran-2-yloxy)-6-(hydroxymethyl)-tetrahydro-2H-pyran-2,3,4-triol::5-{5-[3,4-Dihydroxy-6-methyl-5-(4,5,6-trihydroxy-3-hydroxymethyl-cyclohex-2-enylamino)-tetrahydro-pyran-2-yloxy]-3,4-dihydroxy-6-hydroxymethyl-tetrahydro-pyran-2-yloxy}-6-hydroxymethyl-tetrahydro-pyran-2,3,4-triol::BAY-G 5421::CHEMBL1566::Glucobay::Precose::acarbose
SMILES C[C@H]1O[C@H](O[C@@H]2[C@@H](CO)O[C@H](O[C@@H]3[C@@H](CO)O[C@@H](O)[C@H](O)[C@H]3O)[C@H](O)[C@H]2O)[C@H](O)[C@@H](O)[C@@H]1N[C@H]1C=C(CO)[C@@H](O)[C@H](O)[C@H]1O
InChI Key InChIKey=XUFXOAAUWZOOIT-SXARVLRPSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50333465
Found 3 hits for monomerid = 50333465