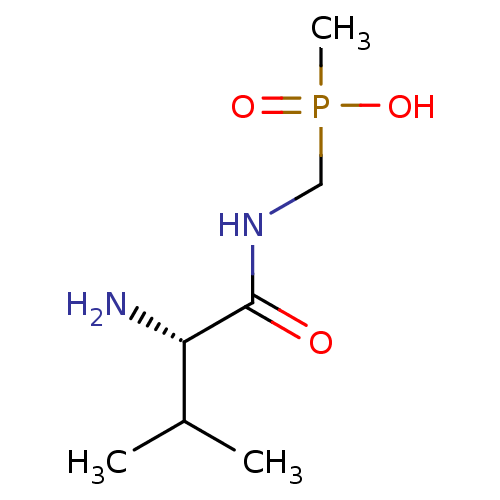
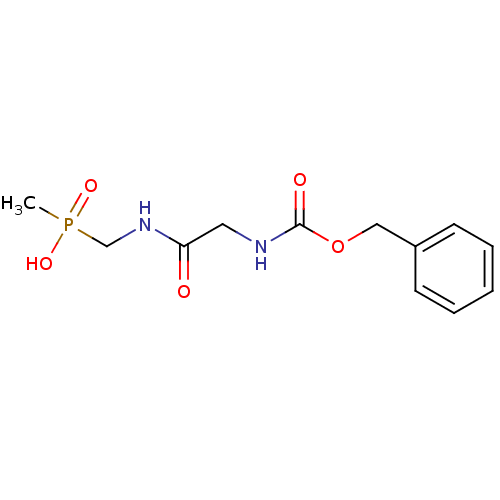
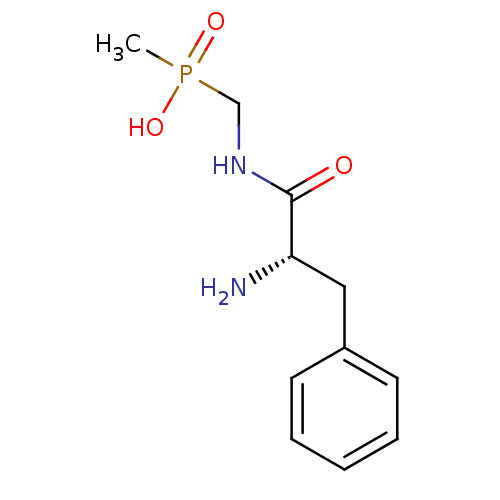
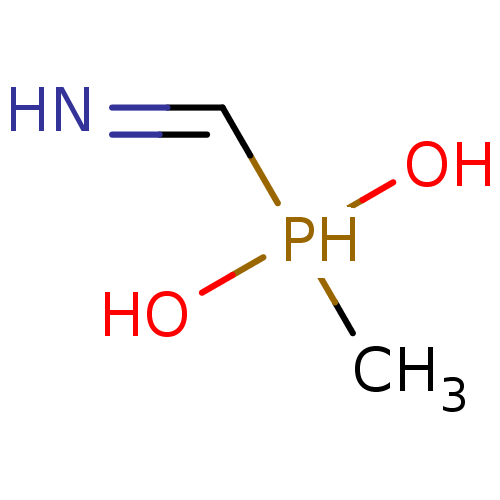
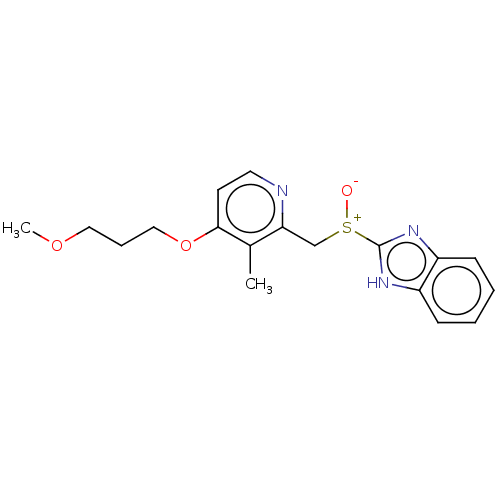
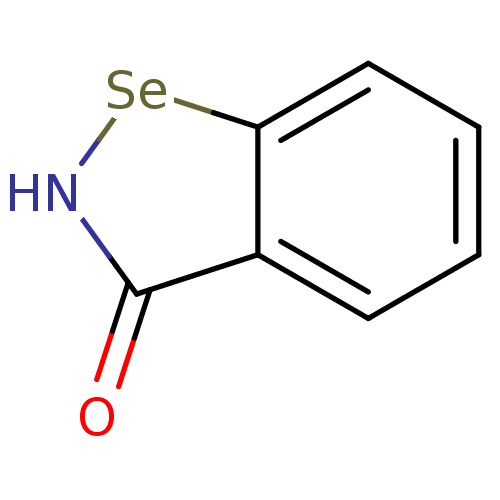
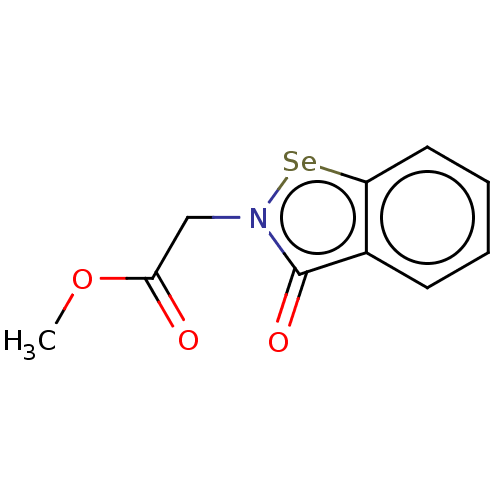
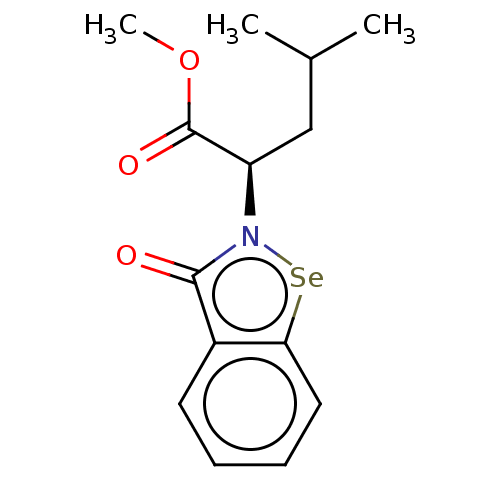
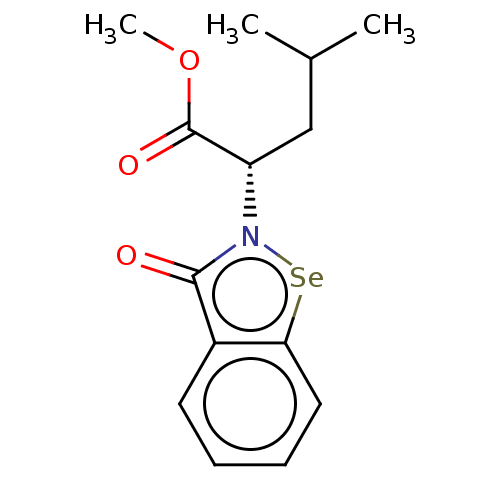
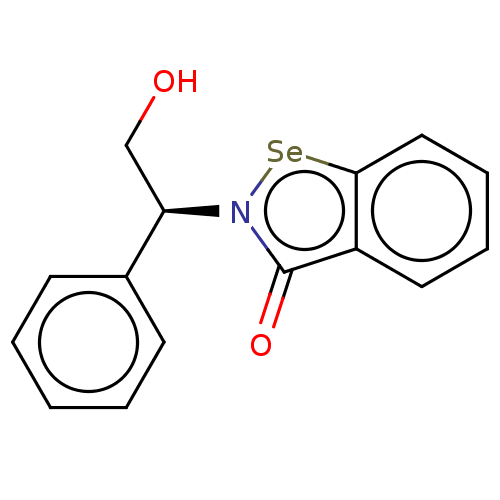
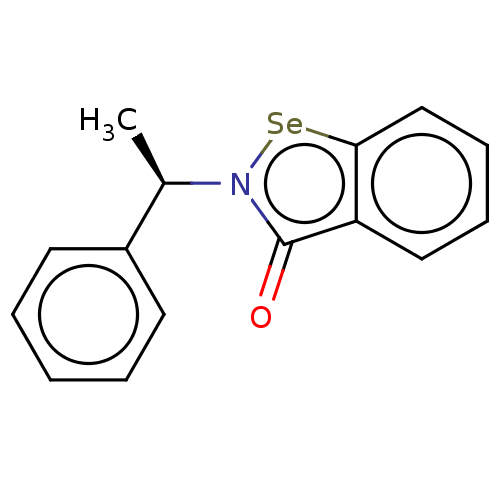
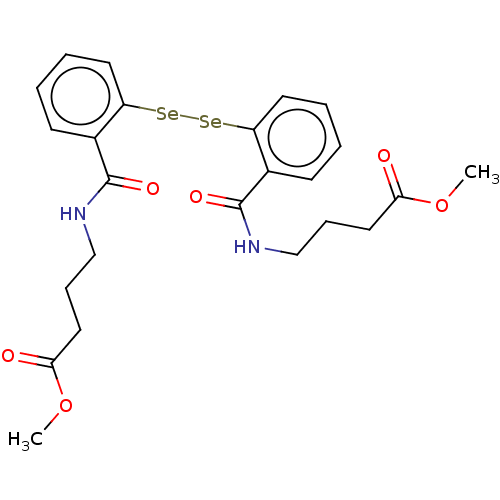
Affinity DataKi: 170nM ΔG°: -39.3kJ/mole IC50: 1.80E+3nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataKi: 184nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataKi: 226nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
Affinity DataKi: 450nM ΔG°: -36.8kJ/mole IC50: 3.10E+3nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataKi: 651nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataKi: 915nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataKi: 3.26E+3nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
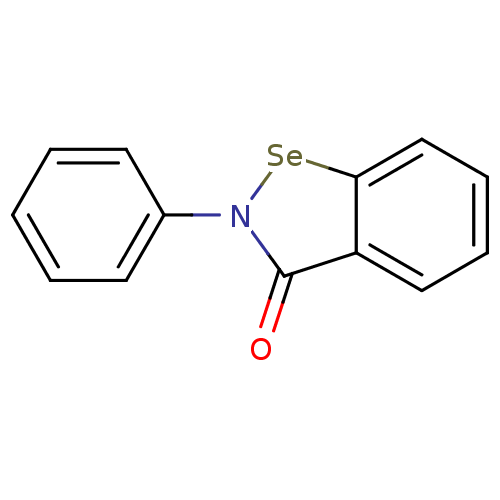
Affinity DataKi: 1.40E+4nM ΔG°: -28.2kJ/mole IC50: 1.58E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 1.75E+4nM ΔG°: -27.6kJ/mole IC50: 1.12E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM ΔG°: -27.5kJ/mole IC50: 6.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 2.10E+4nM ΔG°: -27.1kJ/mole IC50: 6.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 2.50E+4nM ΔG°: -26.7kJ/mole IC50: 8.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+4nM ΔG°: -26.5kJ/mole IC50: 1.53E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 3.00E+4nM ΔG°: -26.2kJ/mole IC50: 8.60E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 3.26E+4nM ΔG°: -26.0kJ/mole IC50: 1.83E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 3.70E+4nM ΔG°: -25.7kJ/mole IC50: 1.50E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 4.10E+4nM ΔG°: -25.5kJ/mole IC50: 3.42E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 4.30E+4nM ΔG°: -25.3kJ/mole IC50: 1.23E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 6.50E+4nM ΔG°: -24.3kJ/mole IC50: 3.19E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
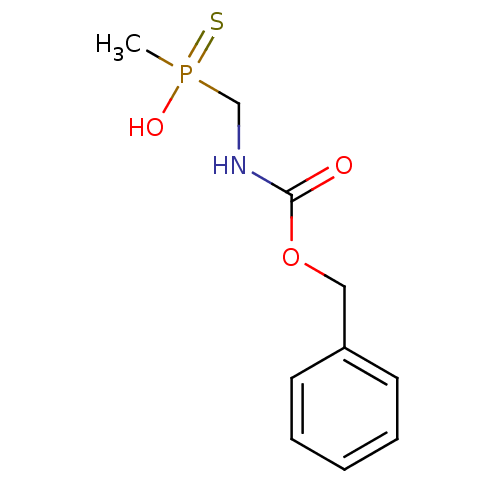
Affinity DataKi: 9.60E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 after 120 mins by Berthelot colorimetric analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.20E+5nM ΔG°: -22.8kJ/mole IC50: 4.85E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 1.35E+5nM ΔG°: -22.5kJ/mole IC50: 4.50E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 1.76E+5nM ΔG°: -21.8kJ/mole IC50: 7.54E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 1.78E+5nM ΔG°: -21.8kJ/mole IC50: 6.40E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 2.08E+5nM ΔG°: -21.4kJ/mole IC50: 6.17E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 2.15E+5nM ΔG°: -21.3kJ/mole IC50: 6.00E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 3.40E+5nM ΔG°: -20.1kJ/mole IC50: 1.10E+6nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataKi: 4.25E+5nM ΔG°: -19.6kJ/mole IC50: 2.53E+6nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
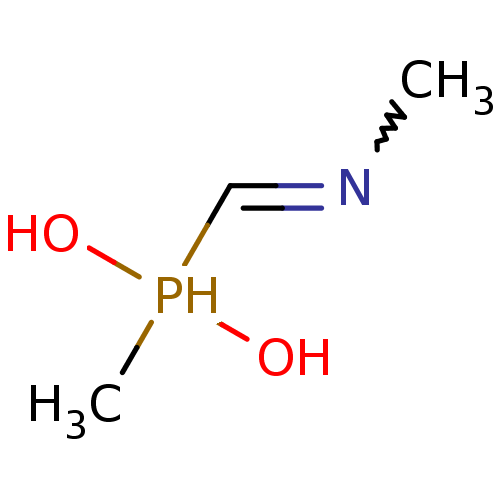
Affinity DataIC50: 290nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as amount of ammonia after 15 mins by indophenol methodMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.28E+3nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 2.12E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.14E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.30E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.47E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate after 2 hrs by Berthelot assayMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.69E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 3.91E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 4.07E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 5.03E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 5.04E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 6.70E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 6.92E+3nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 9.20E+3nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.09E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.18E+4nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 1.90E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 2.01E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 2.27E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 2.53E+4nMAssay Description:Inhibition of recombinant Helicobacter pylori urease expressed in Escherichia coli rosetta DE3 using urea as substrate assessed as transformed cell u...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Wroc?Aw University Of Technology
Curated by ChEMBL
Wroc?Aw University Of Technology
Curated by ChEMBL
Affinity DataIC50: 2.54E+4nMAssay Description:Inhibition of urease in Helicobacter pylori J99 in Brucella broth using urea as substrate preincubated for 2 hrs followed by substrate addition measu...More data for this Ligand-Target Pair

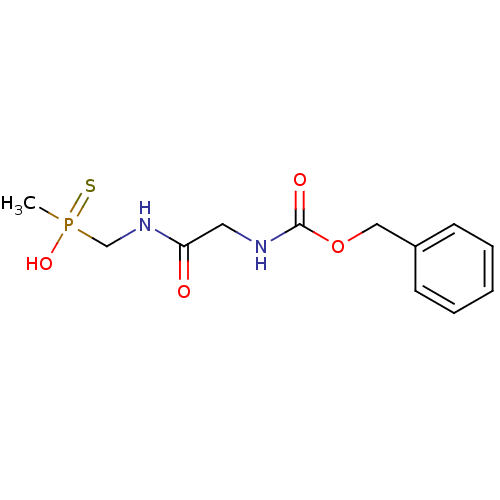



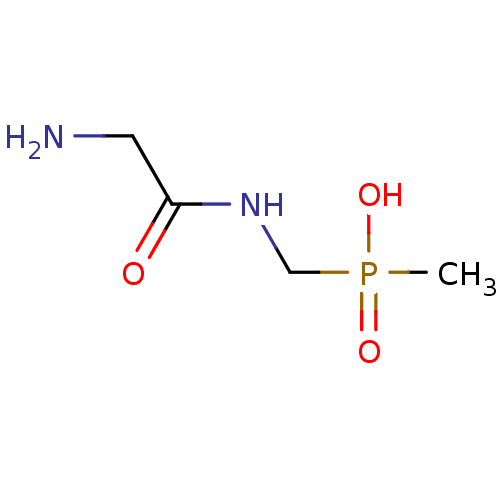
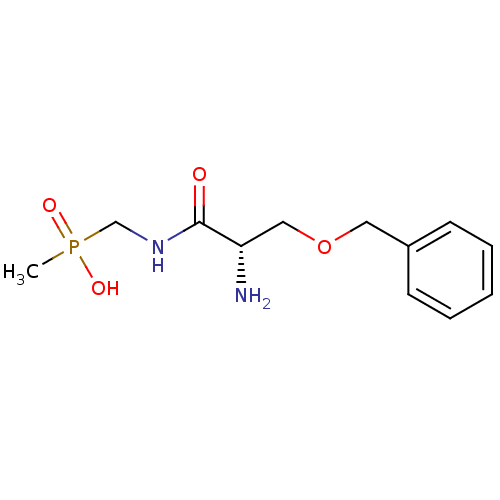
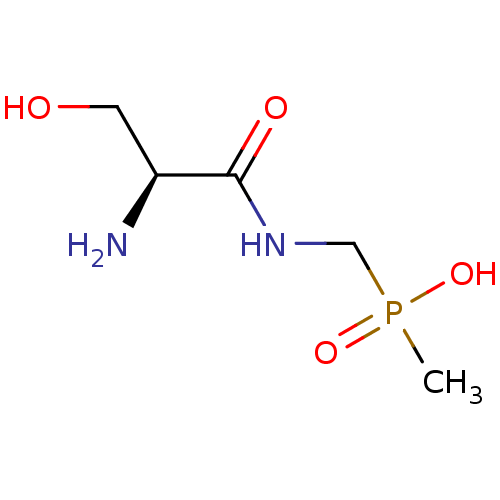
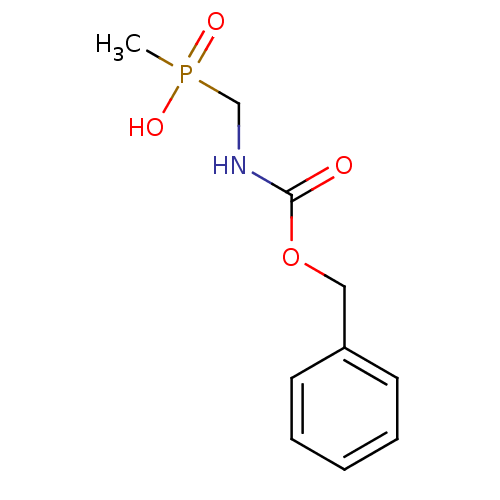
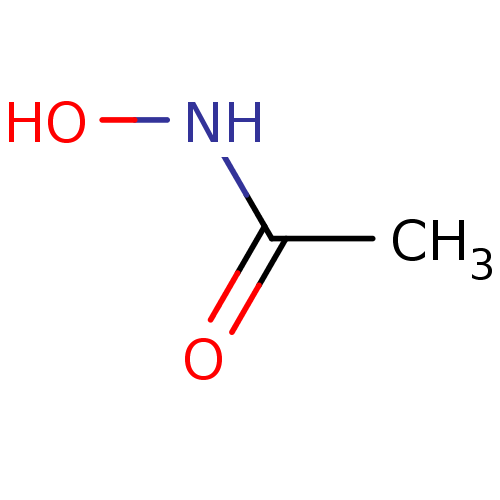
 3D Structure (crystal)
3D Structure (crystal)